SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control - PubMed (original) (raw)
SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control
Shuying Sun et al. Nat Struct Mol Biol. 2010 Mar.
Abstract
SF2/ASF is a prototypical serine- and arginine-rich protein, with important roles in splicing and other aspects of mRNA metabolism. Splicing factor, arginine/serine-rich 1 (SFRS1), the gene encoding SF2/ASF, is a potent proto-oncogene with abnormal expression in many tumors. We found that SF2/ASF negatively autoregulates its expression to maintain homeostatic levels. We characterized six alternatively spliced SF2/ASF mRNA isoforms: the major isoform encodes full-length protein, whereas the others are either retained in the nucleus or degraded by nonsense-mediated mRNA decay. Unproductive splicing accounts for only part of the autoregulation, which occurs primarily at the translational level. The effect is specific to SF2/ASF and requires RNA recognition motif 2 (RRM2). The ultraconserved 3' untranslated region (UTR) is necessary and sufficient for downregulation. SF2/ASF overexpression shifts the distribution of target mRNA toward monoribosomes, and translational repression is partly independent of Dicer and a 5' cap. Thus, multiple post-transcriptional and translational mechanisms are involved in fine-tuning the expression of SF2/ASF.
Figures
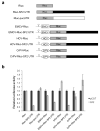
Figure 1
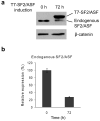
HeLa tet-off cells with inducible SF2/ASF overexpression. (a) Western blot analysis of SF2/ASF before and after induction, using an antibody that recognizes both endogenous and epitope-tagged SF2/ASF. (b) Quantification of endogenous SF2/ASF protein before and after induction. Error bars show standard deviations (SD); n = 3.
Figure 2
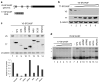
Alternative splicing of SF2/ASF. (a) Six alternative splicing isoforms were identified by RT-PCR with primers a and c, followed by cloning and sequencing. Their structures are shown in the diagrams, with the genomic scale shown at the top. The primers are indicated by arrows below the isoform diagrams. Grey represents protein-coding regions; black denotes the UTRs. The two bars below the genomic scale represent the positions of UCRs-. The correspondence between exon sequences and the domain structure of the protein—including two RNA-recognitions motifs (RRM) and an arginine/serine-rich (RS) domain—is shown at the bottom of the panel. (b) Cycloheximide treatment was used to inhibit NMD. Radioactive RT-PCR with primers b and c was performed to detect the changes of all the alternative splicing isoforms. (c) Cell fractionation was performed to separate nucleus and cytoplasm. RNA from cells before fractionation and from nuclear and cytoplasmic fractions was extracted for radioactive RT-PCR with primers b and c. T: total; N: nucleus; C: cytoplasm. (d) RT-PCR of RNAs from the same cell samples as in Figure 1, before and after SF2/ASF induction. The bottom panel shows amplification of a region common to all the isoforms, using primers d and e. (e) Quantification of the SF2/ASF protein-coding mRNA, isoform I, before and after induction. Error bars show SD; n = 3. t-test, P < 0.04
Figure 3
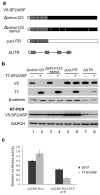
Expression of SF2/ASF from a genomic construct. (a) Diagrams of the V5-tagged SF2/ASF genomic construct and T7-tagged SF2/ASF cDNA construct. The V5 epitope tag is indicated by an open circle, and the T7 tag by an open hexagon. RT-PCR primers used in panel d are indicated by arrows. (b) V5-tagged genomic SF2/ASF was co-transfected with increasing amounts of T7-SF2/ASF cDNA into HeLa cells. After 48 h, protein and RNA were isolated, and Western blotting was performed with both V5 and T7 antibodies, with β-catenin as a normalization control. (c) Genomic V5-SF2/ASF was co-transfected with various T7-tagged SF2/ASF mutants and other SR protein cDNAs. Western blotting was performed with both V5 and T7 antibodies. The histogram shows the quantification of the relative V5-SF2/ASF expression level. The level of V5-tagged SF2/ASF was measured and normalized to that of each T7-tagged protein, with wild-type T7-SF2/ASF as the standard. The level of V5-SF2/ASF co-transfected with empty vector was set at 1. (d) RT-PCR of V5-SF2/ASF with one primer in the V5 tag and the other in SF2/ASF exon 3. The band corresponding to spliced mRNA is indicated by an arrow. *, RNAs that retained one or more introns.
Figure 4
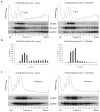
The 3′UTR is necessary and sufficient for SF2/ASF autoregulation. (a) Diagrams of the genomic V5-SF2/ASF mutants. Grey represents the coding region; black represents the natural 3′UTR; white represents a heterologous sequence of the same length; and the thin light-grey bar represents the 3′UTR sequences from the vector; the V5 tag is denoted by an open circle; the grey vertical lines in the 3′UTR represent inactivating mutations of the alternative 5′ and 3′ splice sites. (b) HeLa cells were co-transfected with V5-SF2/ASF genomic mutants and T7-SF2/ASF cDNA. Western blotting was performed to detect SF2/ASF expressed from the genomic construct using V5 antibody, from the cDNA using T7 antibody, and endogenous β-catenin was detected as a loading control. RT-PCR was carried out using the same primers as in Fig. 3d, with GAPDH as a reference. Deletion of the 3′UTR results in much more efficient translation, so in lanes 7 and 8 we transfected only 1/5 as much reporter plasmid, and loaded 1/2 as much total protein. (c) Luciferase reporter assay. The 3′UTR of SF2/ASF was fused to a Renilla luciferase reporter gene. The reporter was co-transfected into HeLa cells with control vector or SF2/ASF cDNA. Luciferase activity was measured and normalized to the luciferase mRNA level determined by radioactive RT-PCR. The relative luciferase activity of pcDNA-Rluc in the absence of SF2/ASF was set at 100%. The error bars show SD; n = 3. t-test, P<0.0001.
Figure 5
SF2/ASF reduces the polysome association of its own mRNA. (a) Sucrose-gradient fractionation of cytoplamic extracts from HeLa cells expressing V5-SF2/ASF Δintron123, Rluc-pucUTR, with (right panel) or without (left panel) T7-SF2/ASF cDNA. Top, UV absorbance (254 nm) profile. Middle and bottom panels, RNA extracted from each fraction was analyzed by radioactive RT-PCR to amplify V5-SF2/ASF, Rluc-pucUTR, and endogenous GAPDH mRNAs. (b) Quantitation of V5-SF2/ASF mRNA distribution in polysome gradients. Relative mRNA levels in each fraction were calculated as percentage of the total levels from all the fractions. Error bars indicate SD; n = 3. (c) Transfected HeLa cells were treated with puromycin for 1 h prior to lysis and fractionation. Gradient fractionation and analysis were done as in a.
Figure 6
IRES-dependent translation assay. (a) Diagrams of EMCV, HCV, and CrPV IRES Renilla-luciferase reporter constructs, with or without the SF2/ASF 3′UTR (black box) or a heterologous sequence of the same length (white box). A hairpin was placed upstream of each IRES to inhibit cap-dependent translation. (b) Luciferase assay. The various Renilla-luciferase reporter constructs were co-transfected into HeLa cells with empty vector or T7-SF2/ASF cDNA. Luciferase activity was normalized to the luciferase mRNA level, measured by radioactive RT-PCR, as in Fig. 4c, and the percent change in the presence of SF2/ASF, compared to the activity in the absence of SF2/ASF, was plotted. The error bars show SD; n = 3.
Similar articles
- The gene encoding the splicing factor SF2/ASF is a proto-oncogene.
Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. Karni R, et al. Nat Struct Mol Biol. 2007 Mar;14(3):185-93. doi: 10.1038/nsmb1209. Epub 2007 Feb 18. Nat Struct Mol Biol. 2007. PMID: 17310252 Free PMC article. - Alternative splicing factor or splicing factor-2 plays a key role in intron retention of the endoglin gene during endothelial senescence.
Blanco FJ, Bernabeu C. Blanco FJ, et al. Aging Cell. 2011 Oct;10(5):896-907. doi: 10.1111/j.1474-9726.2011.00727.x. Epub 2011 Jul 19. Aging Cell. 2011. PMID: 21668763 - MicroRNA (miRNA)-mediated interaction between leukemia/lymphoma-related factor (LRF) and alternative splicing factor/splicing factor 2 (ASF/SF2) affects mouse embryonic fibroblast senescence and apoptosis.
Verduci L, Simili M, Rizzo M, Mercatanti A, Evangelista M, Mariani L, Rainaldi G, Pitto L. Verduci L, et al. J Biol Chem. 2010 Dec 10;285(50):39551-63. doi: 10.1074/jbc.M110.114736. Epub 2010 Oct 4. J Biol Chem. 2010. PMID: 20923760 Free PMC article. - Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins.
Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R. Lemaire R, et al. Genes Dev. 2002 Mar 1;16(5):594-607. doi: 10.1101/gad.939502. Genes Dev. 2002. PMID: 11877379 Free PMC article. - Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF.
Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF. Sanford JR, et al. PLoS One. 2008 Oct 8;3(10):e3369. doi: 10.1371/journal.pone.0003369. PLoS One. 2008. PMID: 18841201 Free PMC article.
Cited by
- Correlation of SRSF1 and PRMT1 expression with clinical status of pediatric acute lymphoblastic leukemia.
Zou L, Zhang H, Du C, Liu X, Zhu S, Zhang W, Li Z, Gao C, Zhao X, Mei M, Bao S, Zheng H. Zou L, et al. J Hematol Oncol. 2012 Jul 27;5:42. doi: 10.1186/1756-8722-5-42. J Hematol Oncol. 2012. PMID: 22839530 Free PMC article. - Regulated Intron Retention and Nuclear Pre-mRNA Decay Contribute to PABPN1 Autoregulation.
Bergeron D, Pal G, Beaulieu YB, Chabot B, Bachand F. Bergeron D, et al. Mol Cell Biol. 2015 Jul;35(14):2503-17. doi: 10.1128/MCB.00070-15. Epub 2015 May 11. Mol Cell Biol. 2015. PMID: 25963658 Free PMC article. - TDP-43 regulates its mRNA levels through a negative feedback loop.
Ayala YM, De Conti L, Avendaño-Vázquez SE, Dhir A, Romano M, D'Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, Baralle FE. Ayala YM, et al. EMBO J. 2011 Jan 19;30(2):277-88. doi: 10.1038/emboj.2010.310. Epub 2010 Dec 3. EMBO J. 2011. PMID: 21131904 Free PMC article. - The translational landscape of the splicing factor SRSF1 and its role in mitosis.
Maslon MM, Heras SR, Bellora N, Eyras E, Cáceres JF. Maslon MM, et al. Elife. 2014 May 6;3:e02028. doi: 10.7554/eLife.02028. Online ahead of print. Elife. 2014. PMID: 24842991 Free PMC article. - Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function.
Haward F, Maslon MM, Yeyati PL, Bellora N, Hansen JN, Aitken S, Lawson J, von Kriegsheim A, Wachten D, Mill P, Adams IR, Caceres JF. Haward F, et al. Elife. 2021 Aug 2;10:e65104. doi: 10.7554/eLife.65104. Elife. 2021. PMID: 34338635 Free PMC article.
References
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–98. - PubMed
- Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. - PubMed
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–43. - PubMed
- Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem. 2004;279:31745–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous