Chemical phylogenetics of histone deacetylases - PubMed (original) (raw)
Chemical phylogenetics of histone deacetylases
James E Bradner et al. Nat Chem Biol. 2010 Mar.
Abstract
The broad study of histone deacetylases in chemistry, biology and medicine relies on tool compounds to derive mechanistic insights. A phylogenetic analysis of class I and II histone deacetylases (HDACs) as targets of a comprehensive, structurally diverse panel of inhibitors revealed unexpected isoform selectivity even among compounds widely perceived as nonselective. The synthesis and study of a focused library of cinnamic hydroxamates allowed the identification of, to our knowledge, the first nonselective HDAC inhibitor. These data will guide a more informed use of HDAC inhibitors as chemical probes and therapeutic agents.
Figures
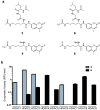
Figure 1
Development of a platform for biochemical profiling of human deacetylases. (a) Chemical structure of substrates 3 - 6. (b) Comparative enzymatic activity of HDAC1-9 with tripeptide substrate 3 and trifluoro acetyl-lysine tripeptide substrate 6, studied at equivalent substrate concentration (10 μM). Data represent mean values of three measurements ± s.d. Substrate 6 allows miniaturized, kinetic study of HDAC4, 5, 7, 8 and 9.
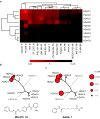
Figure 2
Chemical phylogenetic analysis of HDACs identifies unexpected selectivity of HDAC inhibitors. (a) Hierarchical clustering of HDACs and a representative panel of structurally-diverse HDAC inhibitor tool and investigational compounds 1, 2, 7-20 weighted by inhibitory potency (Ki). Complete quantitative data are shown in Supplementary Table 1. Data based on mean values of triplicate measurements. (b,c) Chemical structure and enzymatic selectivity profile of (b) MS-275 19 and (c) SAHA 1, overlaying molecular phylogeny. HDAC dendrograms are adapted from Supplementary Figure 4. Circles are proportionate in size to Ki on a logarithmic scale, as shown.
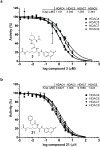
Figure 3
Inhibition of Class IIa HDACs by acetylated lysine based substrates. Inhibition of trifluoroacetyl-lysine substrate 6 processing by (a) acetyl-lysine substrate 3 (b) acetyl-lysine substrate 21 is presented in dose-response format for Class IIa HDACs. Data represent mean values of triplicate measurements ± s.d. IC50 curves were fit by logistic regression.
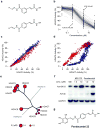
Figure 4
Synthesis and testing of an HDAC-biased chemical library and identification of a non-selective HDAC inhibitor. (a) Library design of meta- and para-substituted hydroxamic acid HDAC inhibitors, utilizing parallel condensation of aldehydes, efficiently samples chemical diversity at the capping feature. (b) Biochemical profiling data for the para-substituted sub-library (n=160 compounds), presented in dose-response format for inhibition of HDAC5. Structural variation in the capping feature was observed to confer a broad range of potency, as illustrated with the most (IC50= 18 nM) and least (IC50 = 55 μM) potent small molecules tested. (c) Comparative biochemical profiling of meta- (red) and para-substituted (blue) sub-libraries for relative inhibition of HDAC2 and HDAC3. The complete library was studied and is displayed at a range of concentrations (0.03, 0.3, 3.0 and 30.0 μM). Compounds of this structural class do not discriminate between HDAC2 and HDAC3. (d) Comparative biochemical profiling of meta- (red) and para-substituted (blue) sub-libraries for relative inhibition of HDAC5 and HDAC7. The complete library was studied and is displayed at a range of concentrations (0.03, 0.3, 3.0 and 30.0 μM). Para-substituted cinnamic hydroxamic acids exhibit increased potency for HDAC5, relative to meta-substituted regioisomers. (e) Specificity profile of pandacostat 22 overlaying molecular phylogeny. HDAC dendrograms are adapted from Supplementary Figure 4. Circles are proportionate in size to Ki on a logarithmic scale, as shown. (f) Immunoblot of Jurkat cells treated with pandacostat for 24 hours and stained for acetylated histones (AcH3K18), acetylated alpha-tubulin (AcTub) or GAPDH. (g) Chemical structure of pandacostat 22.
Similar articles
- Structure-Based Inhibitor Discovery of Class I Histone Deacetylases (HDACs).
Luo Y, Li H. Luo Y, et al. Int J Mol Sci. 2020 Nov 22;21(22):8828. doi: 10.3390/ijms21228828. Int J Mol Sci. 2020. PMID: 33266366 Free PMC article. Review. - Exploration of the binding pocket of histone deacetylases: the design of potent and isoform-selective inhibitors.
Uba Aİ, Yelekçi K. Uba Aİ, et al. Turk J Biol. 2017 Dec 18;41(6):901-918. doi: 10.3906/biy-1701-26. eCollection 2017. Turk J Biol. 2017. PMID: 30814855 Free PMC article. - Zinc-dependent histone deacetylases: Potential therapeutic targets for arterial hypertension.
Kee HJ, Kim I, Jeong MH. Kee HJ, et al. Biochem Pharmacol. 2022 Aug;202:115111. doi: 10.1016/j.bcp.2022.115111. Epub 2022 May 28. Biochem Pharmacol. 2022. PMID: 35640713 Review. - Chemical Versatility in Catalysis and Inhibition of the Class IIb Histone Deacetylases.
Christianson DW. Christianson DW. Acc Chem Res. 2024 Apr 16;57(8):1135-1148. doi: 10.1021/acs.accounts.3c00801. Epub 2024 Mar 26. Acc Chem Res. 2024. PMID: 38530703 Review.
Cited by
- Inhibition of class I histone deacetylase activity represses matrix metalloproteinase-2 and -9 expression and preserves LV function postmyocardial infarction.
Mani SK, Kern CB, Kimbrough D, Addy B, Kasiganesan H, Rivers WT, Patel RK, Chou JC, Spinale FG, Mukherjee R, Menick DR. Mani SK, et al. Am J Physiol Heart Circ Physiol. 2015 Jun 1;308(11):H1391-401. doi: 10.1152/ajpheart.00390.2014. Epub 2015 Mar 20. Am J Physiol Heart Circ Physiol. 2015. PMID: 25795711 Free PMC article. - HDAC6 regulates glucocorticoid receptor signaling in serotonin pathways with critical impact on stress resilience.
Espallergues J, Teegarden SL, Veerakumar A, Boulden J, Challis C, Jochems J, Chan M, Petersen T, Deneris E, Matthias P, Hahn CG, Lucki I, Beck SG, Berton O. Espallergues J, et al. J Neurosci. 2012 Mar 28;32(13):4400-16. doi: 10.1523/JNEUROSCI.5634-11.2012. J Neurosci. 2012. PMID: 22457490 Free PMC article. - Molecular mechanisms of bifunctional vitamin D receptor agonist-histone deacetylase inhibitor hybrid molecules in triple-negative breast cancer.
Barbier C, Mansour A, Ismailova A, Sarmadi F, Scarlata DA, Bouttier M, Zeitouni C, Wang C, Gleason JL, White JH. Barbier C, et al. Sci Rep. 2022 Apr 25;12(1):6745. doi: 10.1038/s41598-022-10740-9. Sci Rep. 2022. PMID: 35468986 Free PMC article. - Coumarin-suberoylanilide hydroxamic acid as a fluorescent probe for determining binding affinities and off-rates of histone deacetylase inhibitors.
Singh RK, Mandal T, Balasubramanian N, Cook G, Srivastava DK. Singh RK, et al. Anal Biochem. 2011 Jan 15;408(2):309-15. doi: 10.1016/j.ab.2010.08.040. Epub 2010 Sep 22. Anal Biochem. 2011. PMID: 20816742 Free PMC article. - A large-scale drug screen identifies selective inhibitors of class I HDACs as a potential therapeutic option for SHH medulloblastoma.
Pak E, MacKenzie EL, Zhao X, Pazyra-Murphy MF, Park PMC, Wu L, Shaw DL, Addleson EC, Cayer SS, Lopez BG, Agar NYR, Rubin LL, Qi J, Merk DJ, Segal RA. Pak E, et al. Neuro Oncol. 2019 Sep 6;21(9):1150-1163. doi: 10.1093/neuonc/noz089. Neuro Oncol. 2019. PMID: 31111916 Free PMC article.
References
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. - PubMed
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. - PubMed
- Choudhary C, et al. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science. 2009;325:834–840. - PubMed
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. - PubMed
Grants and funding
- P01 CA078048-08/CA/NCI NIH HHS/United States
- K08 CA128972-02/CA/NCI NIH HHS/United States
- T32 CA079443-08/CA/NCI NIH HHS/United States
- K08 CA128972-02S1/CA/NCI NIH HHS/United States
- K08 CA128972-01A1/CA/NCI NIH HHS/United States
- N01-C0-012400/CA/NCI NIH HHS/United States
- P01 CA078048-09/CA/NCI NIH HHS/United States
- P01 CA078048-10/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 DA028301/DA/NIDA NIH HHS/United States
- N01-C0-012400/CD/ODCDC CDC HHS/United States
- T32 CA079443/CA/NCI NIH HHS/United States
- R01 DA028301-01/DA/NIDA NIH HHS/United States
- K08 CA128972/CA/NCI NIH HHS/United States
- N01CO12400/CA/NCI NIH HHS/United States
- P01 CA078048/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Molecular Biology Databases