A multigene predictor of outcome in glioblastoma - PubMed (original) (raw)
doi: 10.1093/neuonc/nop007. Epub 2009 Oct 20.
Li Zhang, Erik P Sulman, J Matthew McDonald, Nasrin Latif Shooshtari, Andreana Rivera, Sonya Popoff, Catherine L Nutt, David N Louis, J Gregory Cairncross, Mark R Gilbert, Heidi S Phillips, Minesh P Mehta, Arnab Chakravarti, Christopher E Pelloski, Krishna Bhat, Burt G Feuerstein, Robert B Jenkins, Ken Aldape
Affiliations
- PMID: 20150367
- PMCID: PMC2940562
- DOI: 10.1093/neuonc/nop007
A multigene predictor of outcome in glioblastoma
Howard Colman et al. Neuro Oncol. 2010 Jan.
Abstract
Only a subset of patients with newly diagnosed glioblastoma (GBM) exhibit a response to standard therapy. To date, a biomarker panel with predictive power to distinguish treatment sensitive from treatment refractory GBM tumors does not exist. An analysis was performed using GBM microarray data from 4 independent data sets. An examination of the genes consistently associated with patient outcome, revealed a consensus 38-gene survival set. Worse outcome was associated with increased expression of genes associated with mesenchymal differentiation and angiogenesis. Application to formalin fixed-paraffin embedded (FFPE) samples using real-time reverse-transcriptase polymerase chain reaction assays resulted in a 9-gene subset which appeared robust in these samples. This 9-gene set was then validated in an additional independent sample set. Multivariate analysis confirmed that the 9-gene set was an independent predictor of outcome after adjusting for clinical factors and methylation of the methyl-guanine methyltransferase promoter. The 9-gene profile was also positively associated with markers of glioma stem-like cells, including CD133 and nestin. In sum, a multigene predictor of outcome in glioblastoma was identified which appears applicable to routinely processed FFPE samples. The profile has potential clinical application both for optimization of therapy in GBM and for the identification of novel therapies targeting tumors refractory to standard therapy.
Figures
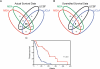
Fig. 1.
Identification of robust outcome-associated genes from microarray data. (A) Overlap of survival genes among 4 microarray data sets. The top 200 genes were identified for each data set individually and the overlap of the 4 lists is shown in a Venn diagram. (B) Estimation of false discovery rate. The survival data were scrambled among the samples and a list of 200 genes was generated from each data set using the scrambled survival data. (C) Survival according to metagene score. The 38 survival-associated genes common to all 4 data sets were used to calculate a metagene score for each sample. The metagene score was calculated by subtracting the sum of the values of the good-prognosis genes from the sum of the values of the poor-prognosis genes. The samples were ranked by metagene score and divided into 2 groups based on results from recursive partitioning analysis. Survival according to metagene score is shown for the group with the lower ranking metagene scores (red) vs samples with higher metagene scores (blue).
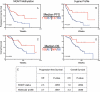
Fig. 2.
Validation of multigene predictor for overall survival in an independent sample set. A set of 68 formalin-fixed, paraffin-embedded glioblastoma samples was subject to qRT-PCR for the 38 gene set identified in Fig. 1. A metagene score was calculated as in Fig. 1 and the samples were ranked by metagene score. Patients were dichotomized into 2 groups based on metagene score using proportions identical to those in Fig. 1. Survival is shown for the lower metagene scores (red) vs the higher metagene scores (blue). Analyses were performed for the entire 38-gene set as well as a smaller 9-gene profile composed of those genes that had the highest individual survival association in the tumors and showed high technical feasibility in paraffin tissues. (A) and (B) Progression-free survival (PFS) according to the entire 38-gene set (A) as well as the 9 gene-profile (B). (C) and (D) Overall survival (OS) according to the 38-gene set (C) and 9-gene set (D). NR, median not reached.
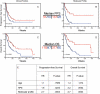
Fig. 3.
Validation of 9-gene profile in temozolomide (TMZ)-treated GBM and comparison with MGMT status. Glioblastoma samples from temozolomide-treated patients (n = 101) were tested for MGMT methylation as well as the 9-gene predictor. For all Kaplan–Meier curves, red indicates low score and blue indicates high score. Tumors were ranked by metagene score and divided into distinct metagene groups using the same cutoffs as in Fig. 1. Progression-free survival (PFS) according to the entire MGMT status (A) as well as the 9-gene profile. (C) and (D) Overall survival (OS) according to MGMT status (C) and 9-gene set (D). (E) Cox proportional hazards multivariate analysis showing that the 9-gene profile is an independent predictor of outcome in TMZ-treated patients after adjusting for MGMT status. NR, median not reached.
Fig. 4.
Comparison of 9-gene molecular profile with clinical variables. Cases were stratified into favorable vs unfavorable clinical groups as descrbed in the text. (A) and (B) Progression-free survival (PFS) according to clinical factors (A), as well as the 9 gene-profile. Median PFS is as shown. (C) and (D) Overall survival (OS) according to MGMT status (C) and 9-gene set (D). Median OS is as shown. (E) Cox proportional hazards multivariate analyses on all (n = 169) validation cases. To generate comparable hazard ratios, patient age was coded as above/below the median and KPS was coded as 70 or above vs <70.
Similar articles
- MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma.
Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, Aldape KD. Rivera AL, et al. Neuro Oncol. 2010 Feb;12(2):116-21. doi: 10.1093/neuonc/nop020. Epub 2009 Dec 14. Neuro Oncol. 2010. PMID: 20150378 Free PMC article. - A five-miRNA signature with prognostic and predictive value for MGMT promoter-methylated glioblastoma patients.
Cheng W, Ren X, Cai J, Zhang C, Li M, Wang K, Liu Y, Han S, Wu A. Cheng W, et al. Oncotarget. 2015 Oct 6;6(30):29285-95. doi: 10.18632/oncotarget.4978. Oncotarget. 2015. PMID: 26320189 Free PMC article. - MiR-106a is an independent prognostic marker in patients with glioblastoma.
Zhao S, Yang G, Mu Y, Han D, Shi C, Chen X, Deng Y, Zhang D, Wang L, Liu Y, Hou X, Wang C, Wu J, Liu H, Wang L, Zhang G, Qi J, Fang X, Shi C, Ai J. Zhao S, et al. Neuro Oncol. 2013 Jun;15(6):707-17. doi: 10.1093/neuonc/not001. Epub 2013 Feb 14. Neuro Oncol. 2013. PMID: 23416698 Free PMC article. - Advances in Experimental Targeted Therapy and Immunotherapy for Patients with Glioblastoma Multiforme.
Polivka J Jr, Polivka J, Holubec L, Kubikova T, Priban V, Hes O, Pivovarcikova K, Treskova I. Polivka J Jr, et al. Anticancer Res. 2017 Jan;37(1):21-33. doi: 10.21873/anticanres.11285. Anticancer Res. 2017. PMID: 28011470 Review.
Cited by
- Sox11 expression in astrocytic gliomas: correlation with nestin/c-Met/IDH1-R132H expression phenotypes, p-Stat-3 and survival.
Korkolopoulou P, Levidou G, El-Habr EA, Adamopoulos C, Fragkou P, Boviatsis E, Themistocleous MS, Petraki K, Vrettakos G, Sakalidou M, Samaras V, Zisakis A, Saetta A, Chatziandreou I, Patsouris E, Piperi C. Korkolopoulou P, et al. Br J Cancer. 2013 May 28;108(10):2142-52. doi: 10.1038/bjc.2013.176. Epub 2013 Apr 25. Br J Cancer. 2013. PMID: 23619925 Free PMC article. - Clinical significance and novel mechanism of action of kallikrein 6 in glioblastoma.
Drucker KL, Paulsen AR, Giannini C, Decker PA, Blaber SI, Blaber M, Uhm JH, O'Neill BP, Jenkins RB, Scarisbrick IA. Drucker KL, et al. Neuro Oncol. 2013 Mar;15(3):305-18. doi: 10.1093/neuonc/nos313. Epub 2013 Jan 10. Neuro Oncol. 2013. PMID: 23307575 Free PMC article. - Oncostatin-M differentially regulates mesenchymal and proneural signature genes in gliomas via STAT3 signaling.
Natesh K, Bhosale D, Desai A, Chandrika G, Pujari R, Jagtap J, Chugh A, Ranade D, Shastry P. Natesh K, et al. Neoplasia. 2015 Feb;17(2):225-37. doi: 10.1016/j.neo.2015.01.001. Neoplasia. 2015. PMID: 25748242 Free PMC article. - Novel approaches for glioblastoma treatment: Focus on tumor heterogeneity, treatment resistance, and computational tools.
Valdebenito S, D'Amico D, Eugenin E. Valdebenito S, et al. Cancer Rep (Hoboken). 2019 Dec;2(6):e1220. doi: 10.1002/cnr2.1220. Epub 2019 Nov 11. Cancer Rep (Hoboken). 2019. PMID: 32729241 Free PMC article. - Biology, genetics and imaging of glial cell tumours.
Walker C, Baborie A, Crooks D, Wilkins S, Jenkinson MD. Walker C, et al. Br J Radiol. 2011 Dec;84 Spec No 2(Spec Iss 2):S90-106. doi: 10.1259/bjr/23430927. Br J Radiol. 2011. PMID: 22433833 Free PMC article. Review.
References
- Kleihues P, Cavenee W WHO. Classification of Tumours: Pathology and Genetics of Tumours of the Nervous System. Lyon: IARC Press; 2000.
- Burton EC, Lamborn KR, Feuerstein BG, et al. Genetic aberrations defined by comparative genomic hybridization distinguish long-term from typical survivors of glioblastoma. Cancer Res. 2002;62:6205–6210. - PubMed
- Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. - PubMed
- Haas-Kogan DA, Prados MD, Lamborn KR, et al. Biomarkers to predict response to epidermal growth factor receptor inhibitors. Cell Cycle. 2005;4:1369–1372. - PubMed
- Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials