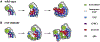dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila - PubMed (original) (raw)
dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila
Mo Weng et al. Dev Cell. 2010.
Abstract
To ensure normal development and maintenance of homeostasis, the extensive developmental potential of stem cells must be functionally distinguished from the limited developmental potential of transit amplifying cells. Yet the mechanisms that restrict the developmental potential of transit amplifying cells are poorly understood. Here we show that the evolutionarily conserved transcription factor dFezf/Earmuff (Erm) functions cell-autonomously to maintain the restricted developmental potential of the intermediate neural progenitors generated by type II neuroblasts in Drosophila larval brains. Although erm mutant intermediate neural progenitors are correctly specified and show normal apical-basal cortical polarity, they can dedifferentiate back into a neuroblast state, functionally indistinguishable from normal type II neuroblasts. Erm restricts the potential of intermediate neural progenitors by activating Prospero to limit proliferation and by antagonizing Notch signaling to prevent dedifferentiation. We conclude that Erm dependence functionally distinguishes intermediate neural progenitors from neuroblasts in the Drosophila larval brain, balancing neurogenesis with stem cell maintenance.
(c) 2010 Elsevier Inc. All rights reserved.
Figures
Figure 1.. erm Mutant Brains Show Ectopic Type II Neuroblasts
(A) A summary of the type II neuroblast lineage. (B-H) While wild-type (+/+) and erm mutant brains contained a similar number of type I neuroblasts (Dpn+CycE+Ase+EdU+; white arrows), erm mutant brains contained ectopic type II neuroblasts (Dpn+CycE+Ase-EdU+; white arrowheads). In (H), wild-type brains contained 85 ± 5.2 type I neuroblasts and 8.0 ± 0 type II neuroblasts, whereas erm mutant brains contained 83.7 ± 6.4 type I neruoblasts and 159 ± 19.7 type II neuroblasts. Scale bar, 20 μm. (I and J) In erm mutant brains expressing GFP driven by Ase-Gal4, Prospero (Pros) always colocalized with Numb (Nb) in metaphase type I neuroblasts (GFP+; white circle), but never in type II neuroblasts (GFP-; white circle). Scale bar, 2 μm. (K and L) erm mutant type I neuroblast clones (white circle) always contained a single neuroblast (white arrow), but erm mutant type II neuroblast clones (white circle) always contained multiple neuroblasts (white arrowheads).
Figure 2.. Erm Maintains the Limited Developmental Potential of INPs
(A and B) At 30 hr after clone induction, both wild-type (+/+) and erm mutant neuroblast clones (yellow circles) contained a single parental neuroblast (white arrows) directly surrounded by immature INPs (white arrowheads) and 1–2 young INPs (Dpn+Ase+). (C-F) At 48 hr after clone induction, wild-type (+/+) neuroblast clones (yellow circles) contained a single parental neuroblast (white arrows) in direct contact with immature INPs (white arrowheads) and young INPs (Dpn+Ase+). Older INPs were away from their parental neuroblasts and were surrounded by GMCs (white asterisks) and neurons(Dpn_Ase_). In contrast, the erm mutant clones contained ectopic type II neuroblast-like cells ([F], yellow arrows) further from the parental neuroblasts than most INPs and neurons. A summary diagram is shown below. (G) R9D11-Gal4 (Erm-Ga14) was undetectable in type II neuroblasts (white arrow) and immature INPs (white arrowheads), but was clearly detected in INPs. All scale bars, 10 μm.
Figure 3.. erm Suppresses the Dedifferentiation of INPs
(A-C) A wild-type (+/+) INP only generated neurons (Dpn_Ase_), but an erm mutant INP generated dedifferentiated neuroblasts (white arrows), immature INPs (white arrowheads) and INPs (Dpn+Ase+), GMCs ([B], white asterisks), and neurons ([C], white asterisks). A lineage clone is circled in yellow, and a summary diagram is shown on the right. (D-I) Similar to wild-type type II neuroblasts, ectopic type II neuroblasts in erm mutant brains lost incorporated EdU (neuroblasts, white arrows; INPs, whitearrow-heads) (D and E), did not express Pros-Gal4 and Erm-Gal4 (type I neuroblast, white arrowheads; type II neuroblasts, white arrows) (F and G), and established ectopic neuroblast lineages (white asterisks) surrounded by glial membrane (H and I). All scale bars, 10 μm.
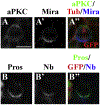
Figure 4.. erm Mutant INPs Show Normal Apical-Basal Polarity
(A and B) Metaphase INPs in erm mutant brains expressing GFP induced by Ase-Gal4 showed asymmetric localization of aPKC, Miranda (Mira), Pros, and Numb (Nb). The scale bar, 5 μm.
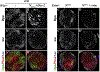
Figure 5.. Erm Restricts the Proliferation of INPs by Promoting Nuclear Prospero
(A) A 3.5 hr pulse of Erm expression induced by Wor-Gal4 was sufficient to trigger Pros localization in neuroblast nuclei (white arrows). (B) Ectopic expression of Erm (57.9 ± 8.6) or Pros (17.4 ± 4.4 neuroblasts) driven by Wor-Gal4 was sufficient to terminate neuroblast proliferation prematurely (98.0 ± 8.4 neuroblasts in wild-type brains). (C and D) Ectopic expression of Erm induced by Erm-Gal4 triggered a significant increased in INPs that exhibited nuclear Pros (white arrows), likely leading them to exit cell cycle prematurely and resulting in some type II neuroblasts (white circle) surrounded only by neurons. Scale bar, 10 μm. (E and F) Overexpression of Pros induced by Erm-Gal4 suppressed ectopic neuroblasts and restored neuronal differentiation in erm mutant brains. Scale bar, 20 μm. (G and H) pros mutant type I neuroblast clones contained ectopic neuroblasts (white arrows). pros mutant type II neuroblast clones contained a single type II neuroblast (white arrow) but showed dramatic overproliferation of INPs (white arrowheads). (I) Overexpression of Erm failed to suppress overproliferation of INPs in pros mutant type II neuroblast clones. Scale bar, 10 μm.
Figure 6.. Erm Suppresses the Dedifferentiation of INPs by Negatively Regulating Notch Signaling
(A and B) Knocking down Notch function by RNAi suppressed ectopic neuroblasts (white arrows) in erm mutant brains. (C and D) Ectopic expression of Erm underthe control of Erm-Gal4 suppressed ectopic neuroblasts induced by constitutive activation of Notch signaling. Scale bar, 20 μm.
Figure 7.. erm Maintains the Restricted Developmental Potential of INPs
(A) Wild-type INPs undergo limited rounds of asymmetric divisions to generate neurons prior to exiting from the cell cycle, and they remain in the same glial chamber as their parental type II neuroblasts. (B) Some erm mutant INPs fail to terminate proliferation and dedifferentiate back into their parental type II neuroblast fate. These dedifferentiated neuroblasts can establish ectopic type II neuroblast lineages and form ectopic glial chambers.
Similar articles
- Earmuff restricts progenitor cell potential by attenuating the competence to respond to self-renewal factors.
Janssens DH, Komori H, Grbac D, Chen K, Koe CT, Wang H, Lee CY. Janssens DH, et al. Development. 2014 Mar;141(5):1036-46. doi: 10.1242/dev.106534. Development. 2014. PMID: 24550111 Free PMC article. - Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development.
Bello BC, Izergina N, Caussinus E, Reichert H. Bello BC, et al. Neural Dev. 2008 Feb 19;3:5. doi: 10.1186/1749-8104-3-5. Neural Dev. 2008. PMID: 18284664 Free PMC article. - The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila.
Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. Bowman SK, et al. Dev Cell. 2008 Apr;14(4):535-46. doi: 10.1016/j.devcel.2008.03.004. Epub 2008 Mar 13. Dev Cell. 2008. PMID: 18342578 Free PMC article. - Diversifying neural cells through order of birth and asymmetry of division.
Zhong W. Zhong W. Neuron. 2003 Jan 9;37(1):11-4. doi: 10.1016/s0896-6273(02)01178-9. Neuron. 2003. PMID: 12526768 Review. - No rest for REST: REST/NRSF regulation of neurogenesis.
Lunyak VV, Rosenfeld MG. Lunyak VV, et al. Cell. 2005 May 20;121(4):499-501. doi: 10.1016/j.cell.2005.05.003. Cell. 2005. PMID: 15907461 Review.
Cited by
- Cortical aPKC kinase activity distinguishes neural stem cells from progenitor cells by ensuring asymmetric segregation of Numb.
Haenfler JM, Kuang C, Lee CY. Haenfler JM, et al. Dev Biol. 2012 May 1;365(1):219-28. doi: 10.1016/j.ydbio.2012.02.027. Epub 2012 Feb 25. Dev Biol. 2012. PMID: 22394487 Free PMC article. - Regulatory modules mediating the complex neural expression patterns of the homeobrain gene during Drosophila brain development.
Hildebrandt K, Kolb D, Klöppel C, Kaspar P, Wittling F, Hartwig O, Federspiel J, Findji I, Walldorf U. Hildebrandt K, et al. Hereditas. 2022 Jan 5;159(1):2. doi: 10.1186/s41065-021-00218-5. Hereditas. 2022. PMID: 34983686 Free PMC article. - Premature translation of the Drosophila zygotic genome activator Zelda is not sufficient to precociously activate gene expression.
Larson ED, Komori H, Fitzpatrick ZA, Krabbenhoft SD, Lee CY, Harrison M. Larson ED, et al. G3 (Bethesda). 2022 Aug 25;12(9):jkac159. doi: 10.1093/g3journal/jkac159. G3 (Bethesda). 2022. PMID: 35876878 Free PMC article. - Fez family transcription factors: controlling neurogenesis and cell fate in the developing mammalian nervous system.
Eckler MJ, Chen B. Eckler MJ, et al. Bioessays. 2014 Aug;36(8):788-97. doi: 10.1002/bies.201400039. Epub 2014 Jun 10. Bioessays. 2014. PMID: 24913420 Free PMC article. Review. - Notch maintains Drosophila type II neuroblasts by suppressing expression of the Fez transcription factor Earmuff.
Li X, Xie Y, Zhu S. Li X, et al. Development. 2016 Jul 15;143(14):2511-21. doi: 10.1242/dev.136184. Epub 2016 May 5. Development. 2016. PMID: 27151950 Free PMC article.
References
- Betschinger J, Mechtler K, and Knoblich JA (2006). Asymmetric segregation of the tumor suppressor Brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253. - PubMed
- Brand AH, and Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous