RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis - PubMed (original) (raw)
RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis
Lin-Yu Lu et al. Dev Cell. 2010.
Abstract
During spermatogenesis, global nucleosome removal occurs where histones are initially replaced by transition proteins and subsequently by protamines. This chromatin reorganization is thought to facilitate the compaction of the paternal genome into the sperm head and to protect the DNA from damaging agents. Histone ubiquitination has been suggested to be important for sex chromosome inactivation during meiotic prophase and nucleosome removal at postmeiotic stages. However, the mechanisms regulating these ubiquitin-mediated processes are unknown. In this study, we investigate the role of the ubiquitin ligase RNF8 during spermatogenesis and find that RNF8-deficient mice are proficient in meiotic sex chromosome inactivation (MSCI) but deficient in global nucleosome removal. Moreover, we show that RNF8-dependent histone ubiquitination induces H4K16 acetylation, which may be an initial step in nucleosome removal. Thus, our results show that RNF8 plays an important role during spermatogenesis through histone ubiquitination, resulting in trans-histone acetylation and global nucleosome removal.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
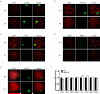
Figure 1. Absence of ubiquitinated H2A in the XY body did not affect meiosis in RNF8-deficient testes
a.-d. Ubiquitinated H2A and ubiquitinated proteins (FK2) were absent in the XY body of RNF8-deficient spermatocytes, while γH2AX and BRCA1 were intact. Typical immunostaining images of spermatocytes at pachytene stage are shown. SCP3 represents synapsed chromosomes and γH2AX marks the XY body. e. RNA polymerase II remained excluded from the XY body in RNF8-deficient spermatocytes. Typical immunostaining images of spermatocytes at pachytene stage are shown. f. Sex chromosome inactivation was maintained in RNF8-deficient mice. Real time PCR results of expression levels of several genes on sex chromosomes in purified spermatocytes are shown. The levels in WT mice were arbitrarily set to 1 and those in RNF8-deficient mice were normalized accordingly. See also Supplementary Figure S3.
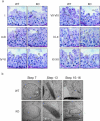
Figure 2. Late steps of spermatid development were abrogated in RNF8-deficient testes
a. RNF8-deficient testes contained less condensed late spermatids. PAS-hematoxylin staining of testes from 3-month old wild type (WT) and RNF8-deficient (KO) mice were performed. Stages of seminiferous epithelium cycle were determined according to the morphology of spermatocytes and round spermatids. Spermatids at different stages are shown. Pl, preleptotene; L, leptotene; Z, zygotene; P, packytene; D, diplotene; RS, round spermatids; ES, elongating spermatids. b. Defect of DNA condensation during step 13-16 of spermatid development was observed in RNF8-deficient testes. Typical electron microscopic images of spermatids at different steps are shown. See also Supplementary Figure S4.
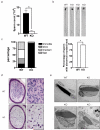
Figure 3. Spermiogenesis was defective in RNF8-deficient mice
a. Number of sperm was decreased in RNF8-deficient mice. Sperm from 3-month old wild type (WT) and RNF8-deficient (KO) mice were counted. Sperm from both cauda epididymis were included. 6 mice of each genotype were used, p<0.001. b. Abnormal rounded sperm heads were observed in RNF8-deficient mice. Upper, typical morphologies of sperm from wild type (WT) and RNF8-deficient (KO) mice with hematoxylin staining are shown; Lower, quantification of sperm with abnormal rounded head is shown. c. Majorities of sperm from RNF8-deficient mice were immotile. Sperm mobility of wild type (WT) and RNF8-deficient (KO) mice are compared. d. H&E staining of paraffin sections of cauda epididymis from wild type (WT) and RNF8-deficient (KO) mice are shown. On right are images with higher magnification showing details of sperm heads. e. Sperm were less condensed and contained residue cytoplasm. Typical electron microscopic images of sperm heads from wild type (WT) and RNF8-deficient (KO) are shown. See also Supplementary Figure S5.
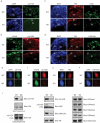
Figure 4. Histones failed to be replaced in sperm from RNF8-deficient mice
a. Protamines failed to replace histones in RNF8-deficient sperm. Typical immunostaining images of sperm from wild type (WT) and RNF8-deficient (KO) mice are shown. Antibodies used are indicated. Prm1: protamine 1; Prm2: protamine 2. b. mRNA levels of transition proteins and protamines were similar in wild type and RNF8-deficient mice. Semi-quantitative RT-PCR was performed using testes cDNA from wild type (WT) and RNF8-deficient (KO) mice. Tnp1: transition protein 1; Tnp2: transition protein 2. GAPDH was used as internal control. c. Incorporation of transition proteins into chromatin was defective in RNF8-deficient mice. Western blots using testes from wild type (WT) and RNF8-deficient (KO) mice are shown. Both NETN-soluble and HCl-released chromatin-bound fractions were used. Antibodies used are indicated, and Plk1 and H3K27me3 were used as soluble and chromatin-bound fraction loading controls respectively. See also Supplementary Figure S6.
Figure 5. Ubiquitinated H2A, H2B and H4 K16 acetylation were decreased in RNF8-deficient testes
a.-d. ub-H2A, ub-H2B and K4 K16 acetylation were decreased in elongating spermatids of RNF8-deficient mice. Typical immunostaining images of testes seminiferous tubules at similar stages from wild type (WT) and RNF8-deficient (KO) mice are shown. SC: spermatocytes; ES: elongating spermatids. Antibodies used are indicated, FK2: antimono/poly-ubiquitinated proteins. e.-h. Typical immunostaining images of individual elongating spermatids from wild type (WT) and RNF8-deficient (KO) mice are shown. i. ub-H2A, ub-H2B and H4 K16 acetylation were decreased in testes from RNF8-deficient mice. Western blots of chromatin-bound proteins using testes from wild type (WT) and RNF8-deficient (KO) mice are shown. Antibodies used are indicated, and H4 was used as loading control.
Figure 6. RNF8 controlled chromatin association of H4K16 acetyltransferase MOF
a. ub-H2A, ub-H2B and H4 K16 acetylation were decreased in RNF8-deficient MEFs. Western blots of chromatin-bound proteins from wild type (WT) and RNF8-deficient (KO) MEFs are shown. Antibodies used are indicated, and H4 was used as loading control. b. Depletion of RNF8 affects not only H2A and H2B ubiquitination, but also H4K16 acetylation. Western blots of chromatin-bound proteins from control siRNA or RNF8 siRNA-treated HeLa cells are shown. c. MOF is enriched in elongating spermatids. Typical immunostaining images of seminiferous tubules at similar stages from wild type (WT) and RNF8-deficient (KO) mice are shown. SC: spermatocytes; ES: elongating spermatids. Antibodies used are indicated. d.-e. Chromatin-associated MOF was decreased in both testes and MEFs from RNF8-defiecient mice. Western blot using wild type (WT) and RNF8-deficient (KO) testes (d) and MEFs (e) are shown. Both NETN-soluble and chromatin-bound fractions were used. Antibodies used are indicated. Plk1 and H3K27me3 were used as soluble and chromatin-bound fraction loading controls, respectively. f. Depletion of MOF affects H4K16 acetylation only, but not H2A and H2B ubiquitination. Western blots of chromatin-bound proteins from control siRNA or MOF siRNA-treated HeLa cells are shown.
Similar articles
- RNF8 and SCML2 cooperate to regulate ubiquitination and H3K27 acetylation for escape gene activation on the sex chromosomes.
Adams SR, Maezawa S, Alavattam KG, Abe H, Sakashita A, Shroder M, Broering TJ, Sroga Rios J, Thomas MA, Lin X, Price CM, Barski A, Andreassen PR, Namekawa SH. Adams SR, et al. PLoS Genet. 2018 Feb 20;14(2):e1007233. doi: 10.1371/journal.pgen.1007233. eCollection 2018 Feb. PLoS Genet. 2018. PMID: 29462142 Free PMC article. - RNF8 is not required for histone-to-protamine exchange in spermiogenesis†.
Abe H, Meduri R, Li Z, Andreassen PR, Namekawa SH. Abe H, et al. Biol Reprod. 2021 Nov 15;105(5):1154-1159. doi: 10.1093/biolre/ioab132. Biol Reprod. 2021. PMID: 34225362 Free PMC article. - Function of RAD6B and RNF8 in spermatogenesis.
Guo Y, Song Y, Guo Z, Hu M, Liu B, Duan H, Wang L, Yuan T, Wang D. Guo Y, et al. Cell Cycle. 2018;17(2):162-173. doi: 10.1080/15384101.2017.1361066. Epub 2018 Jan 19. Cell Cycle. 2018. PMID: 28825854 Free PMC article. - From meiosis to postmeiotic events: the secrets of histone disappearance.
Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S. Gaucher J, et al. FEBS J. 2010 Feb;277(3):599-604. doi: 10.1111/j.1742-4658.2009.07504.x. Epub 2009 Dec 15. FEBS J. 2010. PMID: 20015078 Review. - RNF8-dependent histone ubiquitination during DNA damage response and spermatogenesis.
Ma T, Keller JA, Yu X. Ma T, et al. Acta Biochim Biophys Sin (Shanghai). 2011 May;43(5):339-45. doi: 10.1093/abbs/gmr016. Epub 2011 Mar 28. Acta Biochim Biophys Sin (Shanghai). 2011. PMID: 21444325 Free PMC article. Review.
Cited by
- DDB1- and CUL4-associated factor 8 plays a critical role in spermatogenesis.
Zhang X, Xia Z, Lv X, Li D, Liu M, Zhang R, Ji T, Liu P, Ren R. Zhang X, et al. Front Med. 2021 Apr;15(2):302-312. doi: 10.1007/s11684-021-0851-8. Epub 2021 Apr 14. Front Med. 2021. PMID: 33855678 - Double-strand break repair on sex chromosomes: challenges during male meiotic prophase.
Lu LY, Yu X. Lu LY, et al. Cell Cycle. 2015;14(4):516-25. doi: 10.1080/15384101.2014.998070. Cell Cycle. 2015. PMID: 25565522 Free PMC article. Review. - How mammals pack their sperm: a variant matter.
Boskovic A, Torres-Padilla ME. Boskovic A, et al. Genes Dev. 2013 Aug 1;27(15):1635-9. doi: 10.1101/gad.226167.113. Genes Dev. 2013. PMID: 23913918 Free PMC article. - Regulation of the DNA damage response on male meiotic sex chromosomes.
Lu LY, Xiong Y, Kuang H, Korakavi G, Yu X. Lu LY, et al. Nat Commun. 2013;4:2105. doi: 10.1038/ncomms3105. Nat Commun. 2013. PMID: 23812044 Free PMC article. - RNF8 up-regulates AR/ARV7 action to contribute to advanced prostate cancer progression.
Zhou T, Wang S, Song X, Liu W, Dong F, Huo Y, Zou R, Wang C, Zhang S, Liu W, Sun G, Lin L, Zeng K, Dong X, Guo Q, Yi F, Wang Z, Li X, Jiang B, Cao L, Zhao Y. Zhou T, et al. Cell Death Dis. 2022 Apr 15;13(4):352. doi: 10.1038/s41419-022-04787-9. Cell Death Dis. 2022. PMID: 35428760 Free PMC article.
References
- Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Molecular cell. 2000;5:367–375. - PubMed
- Allfrey VG, Pogo BG, Littau VC, Gershey EL, Mirsky AE. Histone acetylation in insect chromosomes. Science (New York, NY. 1968;159:314–316. - PubMed
- Baarends WM, Hoogerbrugge JW, Roest HP, Ooms M, Vreeburg J, Hoeijmakers JH, Grootegoed JA. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Developmental biology. 1999;207:322–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA132755-04/CA/NCI NIH HHS/United States
- R01 CA132755/CA/NCI NIH HHS/United States
- CA130899/CA/NCI NIH HHS/United States
- R01 CA130899/CA/NCI NIH HHS/United States
- R01 CA130899-03/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials