Practical application of bioinformatics by the multidisciplinary VIZIER consortium - PubMed (original) (raw)
Review
Practical application of bioinformatics by the multidisciplinary VIZIER consortium
Alexander E Gorbalenya et al. Antiviral Res. 2010 Aug.
Abstract
This review focuses on bioinformatics technologies employed by the EU-sponsored multidisciplinary VIZIER consortium (Comparative Structural Genomics of Viral Enzymes Involved in Replication, FP6 PROJECT: 2004-511960, active from 1 November 2004 to 30 April 2009), to achieve its goals. From the management of the information flow of the project, to bioinformatics-mediated selection of RNA viruses and prediction of protein targets, to the analysis of 3D protein structures and antiviral compounds, these technologies provided a communication framework and integrated solutions for steady and timely advancement of the project. RNA viruses form a large class of major pathogens that affect humans and domestic animals. Such RNA viruses as HIV, Influenza virus and Hepatitis C virus are of prime medical concern today, but the identities of viruses that will threaten human population tomorrow are far from certain. To contain outbreaks of common or newly emerging infections, prototype drugs against viruses representing the Virus Universe must be developed. This concept was championed by the VIZIER project which brought together experts in diverse fields to produce a concerted and sustained effort for identifying and validating targets for antivirus therapy in dozens of RNA virus lineages.
Copyright 2010 Elsevier B.V. All rights reserved.
Figures
Fig. 1
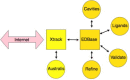
VIZIER targets data pathway. Information stored into the VIZIER Targets Database can be accessed through the three VIZIER Targets DB interfaces including: (A) Targets Data; (B) Targets Status; (C) Target Focus. Xtrack provides also three Interfaces for users that are (D)–(F). Both VIZIER and Xtrack databases are inter-linked. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 2
Rooted phylogenetic analysis of C cluster Human Enteroviruses for the structural part of polyprotein. Figure was modified from Fig. 1 in (Jiang et al., 2007).
Fig. 3
Separate domains and their combinations (targets) predicted for RNA viruses with Viralis.
Fig. 4
The VaZyMolO Browser Focus Interface. It allows browsing from genomic information to a protein's module details.
Fig. 5
The MeDor program's output for phosphoprotein P24 of the Borna disease virus (accession number
P26668
). The highlighted regions in red and blue, and the textual comments constitute annotations made by a user. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 6
Viralis Web interface to handle user queries and access targets predicted in RNA viruses.
Fig. 7
Viralis Domain Prediction Response module interface.
Fig. 8
ID-to-name conversion in tree of alphaviruses by SNAD.
Fig. 9
Xtrack/EDBase components and interactions.
Fig. 10
Xtrack project listing, centred on target VZ93. Some fields are clipped to the right of the image.
Fig. 11
Access to EDBase for VZ93 example. (a) Four models (with goodness-of-fit indicators) are stored in the database and (b) solvent B-factor distribution after refinement.
Fig. 12
(a) Access to ValLigURL for the VZ95 example. (b) JMol superposition of SAM (s-adenosylmethionine) to PDB entry 1JG4.
Similar articles
- The VIZIER project: overview; expectations; and achievements.
Coutard B, Canard B. Coutard B, et al. Antiviral Res. 2010 Aug;87(2):85-94. doi: 10.1016/j.antiviral.2010.02.326. Epub 2010 Mar 10. Antiviral Res. 2010. PMID: 20226212 Free PMC article. Review. - The VIZIER project: preparedness against pathogenic RNA viruses.
Coutard B, Gorbalenya AE, Snijder EJ, Leontovich AM, Poupon A, De Lamballerie X, Charrel R, Gould EA, Gunther S, Norder H, Klempa B, Bourhy H, Rohayem J, L'hermite E, Nordlund P, Stuart DI, Owens RJ, Grimes JM, Tucker PA, Bolognesi M, Mattevi A, Coll M, Jones TA, Aqvist J, Unge T, Hilgenfeld R, Bricogne G, Neyts J, La Colla P, Puerstinger G, Gonzalez JP, Leroy E, Cambillau C, Romette JL, Canard B. Coutard B, et al. Antiviral Res. 2008 Apr;78(1):37-46. doi: 10.1016/j.antiviral.2007.10.013. Epub 2007 Nov 29. Antiviral Res. 2008. PMID: 18083241 Free PMC article. - Understanding the alphaviruses: recent research on important emerging pathogens and progress towards their control.
Gould EA, Coutard B, Malet H, Morin B, Jamal S, Weaver S, Gorbalenya A, Moureau G, Baronti C, Delogu I, Forrester N, Khasnatinov M, Gritsun T, de Lamballerie X, Canard B. Gould EA, et al. Antiviral Res. 2010 Aug;87(2):111-24. doi: 10.1016/j.antiviral.2009.07.007. Epub 2009 Jul 16. Antiviral Res. 2010. PMID: 19616028 Free PMC article. Review. - Genomics and structure/function studies of Rhabdoviridae proteins involved in replication and transcription.
Assenberg R, Delmas O, Morin B, Graham SC, De Lamballerie X, Laubert C, Coutard B, Grimes JM, Neyts J, Owens RJ, Brandt BW, Gorbalenya A, Tucker P, Stuart DI, Canard B, Bourhy H. Assenberg R, et al. Antiviral Res. 2010 Aug;87(2):149-61. doi: 10.1016/j.antiviral.2010.02.322. Epub 2010 Feb 25. Antiviral Res. 2010. PMID: 20188763 Review. - Structure and functionality in flavivirus NS-proteins: perspectives for drug design.
Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, Cook S, Coutard B, Decroly E, de Lamballerie X, Gould EA, Grard G, Grimes JM, Hilgenfeld R, Jansson AM, Malet H, Mancini EJ, Mastrangelo E, Mattevi A, Milani M, Moureau G, Neyts J, Owens RJ, Ren J, Selisko B, Speroni S, Steuber H, Stuart DI, Unge T, Bolognesi M. Bollati M, et al. Antiviral Res. 2010 Aug;87(2):125-48. doi: 10.1016/j.antiviral.2009.11.009. Epub 2009 Nov 27. Antiviral Res. 2010. PMID: 19945487 Free PMC article. Review.
Cited by
- Viral Instant Mutation Viewer: A Tool to Speed Up the Identification and Analysis of New SARS-CoV-2 Emerging Variants and Beyond.
Wilde V, Canard B, Ferron F. Wilde V, et al. Viruses. 2023 Jul 26;15(8):1628. doi: 10.3390/v15081628. Viruses. 2023. PMID: 37631971 Free PMC article. - Genome-wide diversity of Zika virus: Exploring spatio-temporal dynamics to guide a new nomenclature proposal.
Seabra SG, Libin PJK, Theys K, Zhukova A, Potter BI, Nebenzahl-Guimaraes H, Gorbalenya AE, Sidorov IA, Pimentel V, Pingarilho M, de Vasconcelos ATR, Dellicour S, Khouri R, Gascuel O, Vandamme AM, Baele G, Cuypers L, Abecasis AB. Seabra SG, et al. Virus Evol. 2022 Mar 29;8(1):veac029. doi: 10.1093/ve/veac029. eCollection 2022. Virus Evol. 2022. PMID: 35478717 Free PMC article. - Translating genomic exploration of the family Polyomaviridae into confident human polyomavirus detection.
Kamminga S, Sidorov IA, Tadesse M, van der Meijden E, de Brouwer C, Zaaijer HL, Feltkamp MCW, Gorbalenya AE. Kamminga S, et al. iScience. 2021 Dec 11;25(1):103613. doi: 10.1016/j.isci.2021.103613. eCollection 2022 Jan 21. iScience. 2021. PMID: 35036862 Free PMC article. - Coronavirus replication-transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit.
Slanina H, Madhugiri R, Bylapudi G, Schultheiß K, Karl N, Gulyaeva A, Gorbalenya AE, Linne U, Ziebuhr J. Slanina H, et al. Proc Natl Acad Sci U S A. 2021 Feb 9;118(6):e2022310118. doi: 10.1073/pnas.2022310118. Proc Natl Acad Sci U S A. 2021. PMID: 33472860 Free PMC article. - A nidovirus perspective on SARS-CoV-2.
Gulyaeva AA, Gorbalenya AE. Gulyaeva AA, et al. Biochem Biophys Res Commun. 2021 Jan 29;538:24-34. doi: 10.1016/j.bbrc.2020.11.015. Epub 2020 Nov 13. Biochem Biophys Res Commun. 2021. PMID: 33413979 Free PMC article. Review.
References
- Bairoch A., UniProt Consortium, Bougueleret L., Altairac S., Amendolia V., Auchincloss A., Argoud-Puy G., Axelsen K., Baratin D., Blatter M.C., Boeckmann B., Bolleman J., Bollondi L., Boutet E., Quintaje S.B., Breuza L., Bridge A., Decastro E., Ciapina L., Coral D., Coudert E., Cusin I., Delbard G., Dornevil D., Roggli P.D., Duvaud S., Estreicher A., Famiglietti L., Feuermann M., Gehant S., Farriol-Mathis N., Ferro S., Gasteiger E., Gateau A., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Hulo N., James J., Jimenez S., Jungo F., Junker V., Kappler T., Keller G., Lachaize C., Lane-Guermonprez L., Langendijk-Genevaux P., Lara V., Lemercier P., Le Saux V., Lieberherr D., Lima T.D., Mangold V., Martin X., Masson P., Michoud K., Moinat M., Morgat A., Mottaz A., Paesano S., Pedruzzi I., Phan I., Pilbout S., Pillet V., Poux S., Pozzato M., Redaschi N., Reynaud S., Rivoire C., Roechert B., Schneider M., Sigrist C., Sonesson K., Staehli S., Stutz A., Sundaram S., Tognolli M., Verbregue L., Veuthey A.L., Yip L., Zuletta L., Apweiler R., Alam-Faruque Y., Antunes R., Barrell D., Binns D., Bower L., Browne P., Chan W.M., Dimmer E., Eberhardt R., Fedotov A., Foulger R., Garavelli J., Golin R., Horne A., Huntley R., Jacobsen J., Kleen M., Kersey P., Laiho K., Leinonen R., Legge D., Lin Q., Magrane M., Martin M.J., O’Donovan C., Orchard S., O’Rourke J., Patient S., Pruess M., Sitnov A., Stanley E., Corbett M., di Martino G., Donnelly M., Luo J., van Rensburg P., Wu C., Arighi C., Arminski L., Barker W., Chen Y.X., Hu Z.Z., Hua H.K., Huang H.Z., Mazumder R., McGarvey P., Natale D.A., Nikolskaya A., Petrova N., Suzek B.E., Vasudevan S., Vinayaka C.R., Yeh L.S., Zhang J. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Research. 2009;37:D169–D174. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous