Hierarchical coarse-graining strategy for protein-membrane systems to access mesoscopic scales - PubMed (original) (raw)
Hierarchical coarse-graining strategy for protein-membrane systems to access mesoscopic scales
Gary S Ayton et al. Faraday Discuss. 2010.
Abstract
An overall multiscale simulation strategy for large scale coarse-grain simulations of membrane protein systems is presented. The protein is modeled as a heterogeneous elastic network, while the lipids are modeled using the hybrid analytic-systematic (HAS) methodology, where in both cases atomistic level information obtained from molecular dynamics simulation is used to parameterize the model. A feature of this approach is that from the outset liposome length scales are employed in the simulation (i.e., on the order of 1/2 a million lipids plus protein). A route to develop highly coarse-grained models from molecular-scale information is proposed and results for N-BAR domain protein remodeling of a liposome are presented.
Figures
Fig. 1
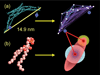
The CG model used in the ED-BAR-liposome simulation. Part (a) shows the hetero-ENM model of the ED-BAR domain as derived from the fully atomistic representation. The two amphipathic helices (highlighted by blue spheres in the left panel) are modeled by two additional sites, as shown in the right panel image. Part (b) depicts the HAS CG lipid model. The fully atomistic lipid is modeled as a single site GB ellipsoid of revolution, augmented with a radially symmetric MS-CG interaction. See ref. for more details. An additional head-group site interacts with the ED-BAR domain, but does not alter the total lipid–lipid interaction.
Fig. 2
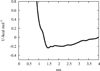
The potential of mean force (PMF) obtained from the distribution function for the distance between the phosphorus atom in the lipid headgroups to the center of mass of the positively charged residues underneath the BAR domain.
Fig. 3
Analysis of the ED-BAR density, mean curvature, H, and relative membrane density change, δρ/ρ0, in the remodeled and isolated liposomes. The original system is a snapshot of the ED-BAR coated liposome. The local ED-BAR density shows local ED-BAR enhancements and depletions that are correlated with both the mean curvature and membrane density. The yellow squares highlight regions where the correlations are most clearly visible. The lower panel on the right shows corresponding plots for the isolated liposome. The scale bar at the bottom gives the reference for each color in the upper panels. The inset at the lower left plots the mean curvature versus liposome density for the two systems. Purple is for the isolated liposome while cyan shows how the local mean curvature is enhanced and distorted because of the ED-BAR membrane remodeling. The upper values of the mean induced curvature correspond to those observed from MD simulations (i.e., about half that observed from MD simulation of a single N-BAR domain, since the mean curvature is the average over two principle curvatures and the single N-BAR domain induces curvature in only one direction).
Fig. 4
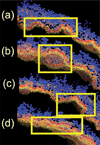
Selected close-up snapshots of the remodeled liposome, obtained by rotating the liposome and selecting out specific regions. The square boxes highlight specific remodeled regions of interest: (a) shows a double hump, (b) a large curved region where the embedded helix sites are visible, (c) a tightly bent region with a high density of ED-BARs on the surface, and (d) a relatively flat region with a correspondingly low ED-BAR density in that region.
Similar articles
- Hybrid coarse-graining approach for lipid bilayers at large length and time scales.
Ayton GS, Voth GA. Ayton GS, et al. J Phys Chem B. 2009 Apr 2;113(13):4413-24. doi: 10.1021/jp8087868. J Phys Chem B. 2009. PMID: 19281167 Free PMC article. - Mesoscopic lateral diffusion in lipid bilayers.
Ayton GS, Voth GA. Ayton GS, et al. Biophys J. 2004 Nov;87(5):3299-311. doi: 10.1529/biophysj.104.047811. Epub 2004 Aug 31. Biophys J. 2004. PMID: 15339807 Free PMC article. - Membrane remodeling from N-BAR domain interactions: insights from multi-scale simulation.
Ayton GS, Blood PD, Voth GA. Ayton GS, et al. Biophys J. 2007 May 15;92(10):3595-602. doi: 10.1529/biophysj.106.101709. Epub 2007 Feb 26. Biophys J. 2007. PMID: 17325001 Free PMC article. - Systematic multiscale simulation of membrane protein systems.
Ayton GS, Voth GA. Ayton GS, et al. Curr Opin Struct Biol. 2009 Apr;19(2):138-44. doi: 10.1016/j.sbi.2009.03.001. Epub 2009 Apr 9. Curr Opin Struct Biol. 2009. PMID: 19362465 Free PMC article. Review. - Coarse-grained simulation: a high-throughput computational approach to membrane proteins.
Sansom MS, Scott KA, Bond PJ. Sansom MS, et al. Biochem Soc Trans. 2008 Feb;36(Pt 1):27-32. doi: 10.1042/BST0360027. Biochem Soc Trans. 2008. PMID: 18208379 Review.
Cited by
- Atomic Modeling of an Immature Retroviral Lattice Using Molecular Dynamics and Mutagenesis.
Goh BC, Perilla JR, England MR, Heyrana KJ, Craven RC, Schulten K. Goh BC, et al. Structure. 2015 Aug 4;23(8):1414-1425. doi: 10.1016/j.str.2015.05.017. Epub 2015 Jun 25. Structure. 2015. PMID: 26118533 Free PMC article. - Long-Range Organization of Membrane-Curving Proteins.
Simunovic M, Šarić A, Henderson JM, Lee KYC, Voth GA. Simunovic M, et al. ACS Cent Sci. 2017 Dec 27;3(12):1246-1253. doi: 10.1021/acscentsci.7b00392. Epub 2017 Nov 21. ACS Cent Sci. 2017. PMID: 29296664 Free PMC article. - Organizing membrane-curving proteins: the emerging dynamical picture.
Simunovic M, Bassereau P, Voth GA. Simunovic M, et al. Curr Opin Struct Biol. 2018 Aug;51:99-105. doi: 10.1016/j.sbi.2018.03.018. Epub 2018 Mar 30. Curr Opin Struct Biol. 2018. PMID: 29609179 Free PMC article. Review. - New insights into BAR domain-induced membrane remodeling.
Ayton GS, Lyman E, Krishna V, Swenson RD, Mim C, Unger VM, Voth GA. Ayton GS, et al. Biophys J. 2009 Sep 16;97(6):1616-25. doi: 10.1016/j.bpj.2009.06.036. Biophys J. 2009. PMID: 19751666 Free PMC article. - Multiscale simulation of protein mediated membrane remodeling.
Ayton GS, Voth GA. Ayton GS, et al. Semin Cell Dev Biol. 2010 Jun;21(4):357-62. doi: 10.1016/j.semcdb.2009.11.011. Epub 2009 Nov 13. Semin Cell Dev Biol. 2010. PMID: 19922811 Free PMC article. Review.
References
- Voth GA, editor. Coarse-graining of condensed phase and biomolecular systems. Boca Raton: CRC Press/Taylor and Francis Group; 2009.
- Singer SJ, Nicolson GL. Science. 1972;175:720–731. - PubMed
- Engelman DM. Nature. 2005;438:578–580. - PubMed
- McLaughlin S, Murray D. Nature. 2005;438:605–611. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources