Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis - PubMed (original) (raw)
Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis
Michael A Kohanski et al. Mol Cell. 2010.
Abstract
Antibiotic resistance arises through mechanisms such as selection of naturally occurring resistant mutants and horizontal gene transfer. Recently, oxidative stress has been implicated as one of the mechanisms whereby bactericidal antibiotics kill bacteria. Here, we show that sublethal levels of bactericidal antibiotics induce mutagenesis, resulting in heterogeneous increases in the minimum inhibitory concentration for a range of antibiotics, irrespective of the drug target. This increase in mutagenesis correlates with an increase in ROS and is prevented by the ROS scavenger thiourea and by anaerobic conditions, indicating that sublethal concentrations of antibiotics induce mutagenesis by stimulating the production of ROS. We demonstrate that these effects can lead to mutant strains that are sensitive to the applied antibiotic but resistant to other antibiotics. This work establishes a radical-based molecular mechanism whereby sublethal levels of antibiotics can lead to multidrug resistance, which has important implications for the widespread use and misuse of antibiotics.
Figures
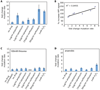
Figure 1. Low levels of bactericidal antibiotics increase mutation rate due to reactive oxygen species formation
(A) Fold change in mutation rate (mean +/− 95% CI) relative to an untreated control (no drug) for wildtype E. coli (MG1655) following an overnight treatment with 1µg/ml ampicillin, 1µg/ml kanamycin, 3µg/ml kanamycin, 15ng/ml norfloxacin, 50ng/ml norfloxacin, or 1mM hydrogen peroxide (H2O2). (B) Correlation between oxidative stress levels (HPF fluorescence) and fold change in mutation rate for wildtype E. coli for the treatments described in A. (C–D) Fold change in mutation rate (mean +/− 95% CI) relative to an untreated control (no drug) for wildtype E. coli following an overnight treatment with 100mM thiourea and no drug, 1µg/ml ampicillin, 1µg/ml kanamycin, 3µg/ml kanamycin, 15ng/ml norfloxacin, 50ng/ml norfloxacin, or 1mM hydrogen peroxide (H2O2) under (C) aerobic growth conditions with 100mM thiourea or (D) anaerobic growth conditions.
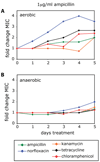
Figure 2. Low levels of bactericidal antibiotics can lead to broad-spectrum increases in MIC due to ROS-mediated mutagenesis
(A–B) Fold change in MIC relative to an aerobic no-drug control for ampicillin, norfloxacin, kanamycin, tetracycline and chloramphenicol, following 5 days of growth in the presence 1µg/ml ampicillin under (A) aerobic or (B) anaerobic growth conditions. See also Figure S1.
Figure 3. Ampicillin treatment of E. coli results in heterogeneous increases in MIC for ampicillin and norfloxacin
(A–B) Shown are the distributions of (A) ampicillin or (B) norfloxacin MICs for 44 ampicillin-treated isolates. The maximum growth inhibitory concentration tested for norfloxacin was 1000ng/ml, and the MICs for these isolates may be ≥ 1000ng/ml.
Figure 4. Ampicillin treatment leads to the formation of norfloxacin-resistant isolates with mutations in gyrA, gyrB or the acrAB promoter (P_acrAB_) and kanamycin-resistant isolates with mutations in rpsL or arcA
(A–B) Isolates with point mutation resulting in (A) a D82G or D87Y substitution in GyrA, or (B) a S464F substitution in GyrB. (C) T-to-A DNA base pair mutation in the AcrR/EnvR binding site of the -35 region of P_acrAB_. P_acrAB_ is partially annotated to show the -10 and -35 regions (bold), the transcription start site (capitalized A) and the AcrR/EnvR binding site (underlined). (D) Isolates with insertion between basepair 92 and 93 (K58) and between basepair 78 and 79 (K62) resulting in truncation of RpsL. (E) Isolate with a single basepair insertion between basepair 211 and 212 resulting in a truncated ArcA protein missing the majority of the helix-turn-helix (HTH) DNA binding domain.
Comment in
- The fast track to multidrug resistance.
Kaufmann BB, Hung DT. Kaufmann BB, et al. Mol Cell. 2010 Feb 12;37(3):297-8. doi: 10.1016/j.molcel.2010.01.027. Mol Cell. 2010. PMID: 20159549 - Reactive resistance.
Molloy S. Molloy S. Nat Rev Genet. 2010 Apr;11(4):240. doi: 10.1038/nrg2769. Nat Rev Genet. 2010. PMID: 21488228 No abstract available.
Similar articles
- The fast track to multidrug resistance.
Kaufmann BB, Hung DT. Kaufmann BB, et al. Mol Cell. 2010 Feb 12;37(3):297-8. doi: 10.1016/j.molcel.2010.01.027. Mol Cell. 2010. PMID: 20159549 - Antidepressant fluoxetine induces multiple antibiotics resistance in Escherichia coli via ROS-mediated mutagenesis.
Jin M, Lu J, Chen Z, Nguyen SH, Mao L, Li J, Yuan Z, Guo J. Jin M, et al. Environ Int. 2018 Nov;120:421-430. doi: 10.1016/j.envint.2018.07.046. Epub 2018 Aug 18. Environ Int. 2018. PMID: 30125859 - Combined exposure to non-antibiotic pharmaceutics and antibiotics in the gut synergistically promote the development of multi-drug-resistance in Escherichia coli.
Shi D, Hao H, Wei Z, Yang D, Yin J, Li H, Chen Z, Yang Z, Chen T, Zhou S, Wu H, Li J, Jin M. Shi D, et al. Gut Microbes. 2022 Jan-Dec;14(1):2018901. doi: 10.1080/19490976.2021.2018901. Gut Microbes. 2022. PMID: 35014598 Free PMC article. - Killing by bactericidal antibiotics does not depend on reactive oxygen species.
Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. Keren I, et al. Science. 2013 Mar 8;339(6124):1213-6. doi: 10.1126/science.1232688. Science. 2013. PMID: 23471410
Cited by
- Defects in DNA double-strand break repair resensitize antibiotic-resistant Escherichia coli to multiple bactericidal antibiotics.
Revitt-Mills SA, Wright EK, Vereker M, O'Flaherty C, McPherson F, Dawson C, van Oijen AM, Robinson A. Revitt-Mills SA, et al. Microbiologyopen. 2022 Oct;11(5):e1316. doi: 10.1002/mbo3.1316. Microbiologyopen. 2022. PMID: 36314749 Free PMC article. - Rapid cytometric antibiotic susceptibility testing utilizing adaptive multidimensional statistical metrics.
Huang TH, Ning X, Wang X, Murthy N, Tzeng YL, Dickson RM. Huang TH, et al. Anal Chem. 2015 Feb 3;87(3):1941-9. doi: 10.1021/ac504241x. Epub 2015 Jan 13. Anal Chem. 2015. PMID: 25540985 Free PMC article. - Global transcriptional responses to the bacteriocin colicin M in Escherichia coli.
Kamenšek S, Žgur-Bertok D. Kamenšek S, et al. BMC Microbiol. 2013 Feb 19;13:42. doi: 10.1186/1471-2180-13-42. BMC Microbiol. 2013. PMID: 23421615 Free PMC article. - Sending out an SOS - the bacterial DNA damage response.
Lima-Noronha MA, Fonseca DLH, Oliveira RS, Freitas RR, Park JH, Galhardo RS. Lima-Noronha MA, et al. Genet Mol Biol. 2022 Oct 10;45(3 Suppl 1):e20220107. doi: 10.1590/1678-4685-GMB-2022-0107. eCollection 2022. Genet Mol Biol. 2022. PMID: 36288458 Free PMC article. - Antibiotic treatment enhances the genome-wide mutation rate of target cells.
Long H, Miller SF, Strauss C, Zhao C, Cheng L, Ye Z, Griffin K, Te R, Lee H, Chen CC, Lynch M. Long H, et al. Proc Natl Acad Sci U S A. 2016 May 3;113(18):E2498-505. doi: 10.1073/pnas.1601208113. Epub 2016 Apr 18. Proc Natl Acad Sci U S A. 2016. PMID: 27091991 Free PMC article.
References
- Andersson DI. Persistence of antibiotic resistant bacteria. Curr Opin Microbiol. 2003;6:452–456. - PubMed
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. - PubMed
- Becnel Boyd L, Maynard MJ, Morgan-Linnell SK, Horton LB, Sucgang R, Hamill RJ, Jimenez JR, Versalovic J, Steffen D, Zechiedrich L. Relationships among ciprofloxacin, gatifloxacin, levofloxacin, and norfloxacin MICs for fluoroquinolone-resistant Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009;53:229–234. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical