Autophagy protects against Sindbis virus infection of the central nervous system - PubMed (original) (raw)
Autophagy protects against Sindbis virus infection of the central nervous system
Anthony Orvedahl et al. Cell Host Microbe. 2010.
Abstract
Autophagy functions in antiviral immunity. However, the ability of endogenous autophagy genes to protect against viral disease in vertebrates remains to be causally established. Here, we report that the autophagy gene Atg5 function is critical for protection against lethal Sindbis virus (SIN) infection of the mouse central nervous system. Inactivating Atg5 in SIN-infected neurons results in delayed clearance of viral proteins, increased accumulation of the cellular p62 adaptor protein, and increased cell death in neurons, but the levels of viral replication remain unaltered. In vitro, p62 interacts with SIN capsid protein, and genetic knockdown of p62 blocks the targeting of viral capsid to autophagosomes. Moreover, p62 or autophagy gene knockdown increases viral capsid accumulation and accelerates virus-induced cell death without affecting virus replication. These results suggest a function for autophagy in mammalian antiviral defense: a cell-autonomous mechanism in which p62 adaptor-mediated autophagic viral protein clearance promotes cell survival.
2010 Elsevier Inc. All rights reserved.
Figures
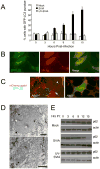
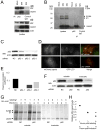
Figure 1. SIN Induces Autophagy in vitro
(A) Quantitation of the percentage of GFP-LC3 MEFs with GFP-LC3 punctae (autophagosomes) after infection with indicated virus. Data shown represent mean ± SEM for triplicate samples of at least 100 cells per sample. Similar results were observed in 3 independent experiments. (B) Representative fluorescent microscopic image demonstrating colocalization of SIN structural proteins (red) with GFP-LC3 (green) in GFP-LC3 MEFs at 12 h post-infection (p.i.). (C) Representative fluorescent microscopic images showing colocalization in Atg5+/+ MEFs or lack of colocalization in _Atg5_−/− MEFs of SIN capsid (red) and GFP-LC3 (green) in cells infected with SIN-mCherry.capsid/GFP-LC3. Images shown represent a single time point at 12 h p.i. from live cell imaging. Arrowhead denotes colocalized puncta; arrow denotes capsid-positive GFP-LC3 ring structure. See movie S1 for dynamic representation of mCherry.capsid and GFP-LC3 localization between 16 and 17 h p.i. in Atg5+/+ MEFs. (D) Representative EMs of wild-type MEFs at 12 h p.i. with SVIA. Left panel demonstrates a double-membraned autophagosome (black arrow) containing SIN nucleocapsids (black arrowheads), cellular membranes (white arrowhead), and aggregates (white arrow). Right panel demonstrates a single-membraned autolysosome with SIN nucleocapsid (black arrowhead). Open arrowheads denote virions budding from the plasma membrane. Scale bars, 200 nm. (E) Measurement of autophagic protein degradation by p62 Western blot analysis in wild-type MEFs at serial time points after mock, SVIA, or UV-SVIA infection.
Figure 2. SIN-Induced Autophagy in Mouse Hippocampal Neurons and Scheme of Experimental Strategies to Inhibit Autophagy in Neurons in vivo
(A) Colocalization of SIN structural proteins (red) with GFP-LC3 in hippocampal neurons of GFP-LC3 transgenic mice 24 h after mock infection (top) or infection with SVIA (bottom). Similar hippocampal regions are shown for mock and SVIA-infected brains. No GFP-LC3 punctae were observed in any regions of the mock-infected brains. (B) Conceptual overview of strategies to inhibit or knock out Atg5 specifically in neurons in vivo, and the relevant control viruses and mouse strains. (i) SIN expressing a dominant negative mutant Atg5 (Atg5K130R). (ii) SIN expressing Cre recombinase in Atg5flox/flox mice. (iii) nestin_-Cre_ mice crossed to Atg5flox/flox mice. (C) Quantitation of the number of GFP-LC3 punctae (autophagosomes) per cell in GFP-LC3 MEFs at 12 h after infection with indicated virus below x axis. Data shown represent mean ± SEM for triplicate samples of at least 100 cells per sample. Similar results were obtained in 3 independent experiments. (D) Detection of genomic Atg5 excision in primary MEFs obtained from Atg5flox/flox mice infected with SIN/Cre or SIN/Cre.Stop.
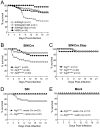
Figure 3. Increased SIN Neurovirulence in Mice with Inactivation of Neuronal Atg5
(A) Mortality of CD1 littermates infected with the indicated recombinant SIN strains or mock-infected with HBSS. Data shown represent combined mortality from 4 independent infections of 8–12 mice per group. Similar results were observed in each independent experiment. (B–C) Mortality of Atg5flox/flox or littermate controls (Atg5+/flox and Atg5+/+) infected with recombinant SIN expressing Cre recombinase or SIN expressing non-coding Cre gene. Data shown in (B–C) represent combined mortality from infection of 22 and 8 separate litters, respectively. (D–E) Mortality of littermates from Atg5+/flox; nestin_-Cre_ transgenic mice crossed with Atg5flox/flox mice and infected with the dsTE12Q strain of SIN (D) or mock-infected (E). Data shown in (D–E) represent combined mortality from infection of 21 and 8 separate litters, respectively.
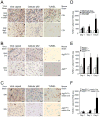
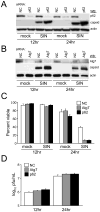
Figure 4. Atg5 Inhibition Delays Viral Antigen Clearance from Neurons Without Affecting SIN CNS Titers
(A–C) SIN titers in mouse brains. Data shown represent geometric mean titers ± SEM for groups of 4–8 mice per time point. (D) SIN antigen staining of brains of mice of the indicated genotype (right labels) infected with the indicated virus (left labels). All micrographs in (D) are from the superior colliculus of the mouse brain, a region that is infected by SIN in all mice in this study. The images shown are representative of the data quantitated in (E–G) for the total mouse brain, with the exception that there is high degree of inter-mouse variability in the level of colliculus staining at day one. Scale bars, 100 μm. (E–G) Quantitation of SIN antigen staining in the brains of mice treated as in (A–C), respectively. Data in (E–G) represent mean number of antigen-positive cells per unit area of mouse brain for 4–8 mice per experimental group.
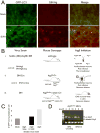
Figure 5. Increased SIN Capsid Staining, Cellular p62 Staining, and Cell Death in the Brains of Mice with Neuronal Atg5 Inactivation
(A–C) Detection of SIN capsid (left column), cellular p62 (middle column), and cell death by TUNEL staining (right column) in mouse brain. Shown are representative photomicrographs for each experimental group of the superior colliculus at day 5 p.i. For (A–C) similar results were observed in 4–8 mice per group. Scale bars, 20 μm. (D–F) Quantitation of number of TUNEL-positive cells per unit area of virus-infected region of brain at days 1, 3, and 5 p.i.. Data in (D–F) represent mean ± SEM for each brain from 4–8 mice per experimental group.
Figure 6. p62 Interacts with SIN Capsid and Targets Capsid for Autophagy
(A–B) Coimmunoprecipitation of SIN capsid with p62 in HeLa/GFP-LC3 cells mock-infected or infected with SVIA (labeled SIN) either by Western blotting with a polyclonal anti-SIN virus capsid antibody (A) or a polyclonal anti-SIN antibody that detects E2/6K, E1, and capsid protein (B). (C–E) Detection of SIN capsid colocalization with GFP-LC3 after p62 knockdown. Western blot analysis of p62 expression in HeLa/GFP-LC3 cells treated with individual p62 siRNA oligos or non-silencing negative control oligos (NC) (C). Representative image of HeLa/GFP-LC3 cells treated with p62-2 siRNA (bottom) or NC (top), and infected with SIN-mCherry.capsid (D). Quantitation of mCherry.capsid colocalization with GFP-LC3 in cells treated with the indicated siRNA (E). Data shown represent the mean of at least 50 infected cells per condition ± SEM. Similar results were obtained in 3 independent experiments. (F–H) Analysis of SIN protein degradation after p62 knockdown. Western blot analysis of p62 expression in the fractions used in (G) for radioimmunoprecipitation (F). Radioimmunoprecipitation with an anti-SIN antibody of soluble and insoluble fractions from HeLa/GPF-LC3 cells treated with NC or p62 siRNA, infected with SVIA, and pulse-chased for the indicated times (G). Quantitation of capsid levels in (G) relative to 1 h control levels for each fraction (H). Closed circles, NC soluble; closed squares, p62 soluble; open circles, NC insoluble; open squares, p62 insoluble. Similar results were observed in 3 independent experiments.
Figure 7. p62 and Atg7 Promote the Survival of SIN-Infected cells
(A–B) Western blot analysis of p62 and capsid expression (A) or Atg7 and capsid expression (B) in HeLa/GFP-LC3 cells treated with the indicated siRNA and infected for the time indicated. (C) Cell death quantitation of cells treated as in (A) and (B) as measured by a trypan blue exclusion assay. Data shown represent mean ± SEM of at least 100 cells per sample for triplicate samples for each condition. (D) Levels of infectious SIN in supernatants of cells in (C). Data represent geometric mean titers ± SEM for triplicate samples. For (A–D), similar results were obtained in three independent experiments.
Comment in
- How autophagy saves mice: A cell-autonomous defense system against Sindbis virus infection.
Yoshimori T. Yoshimori T. Cell Host Microbe. 2010 Feb 18;7(2):83-4. doi: 10.1016/j.chom.2010.02.003. Cell Host Microbe. 2010. PMID: 20159611
Similar articles
- Sindbis Virus Can Exploit a Host Antiviral Protein To Evade Immune Surveillance.
Wang X, Li MMH, Zhao J, Li S, MacDonald MR, Rice CM, Gao X, Gao G. Wang X, et al. J Virol. 2016 Oct 28;90(22):10247-10258. doi: 10.1128/JVI.01487-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581990 Free PMC article. - Germ Line IgM Is Sufficient, but Not Required, for Antibody-Mediated Alphavirus Clearance from the Central Nervous System.
Nilaratanakul V, Chen J, Tran O, Baxter VK, Troisi EM, Yeh JX, Griffin DE. Nilaratanakul V, et al. J Virol. 2018 Mar 14;92(7):e02081-17. doi: 10.1128/JVI.02081-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29321331 Free PMC article. - Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein.
Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Liang XH, et al. J Virol. 1998 Nov;72(11):8586-96. doi: 10.1128/JVI.72.11.8586-8596.1998. J Virol. 1998. PMID: 9765397 Free PMC article. - The role of antibody in recovery from alphavirus encephalitis.
Griffin D, Levine B, Tyor W, Ubol S, Desprès P. Griffin D, et al. Immunol Rev. 1997 Oct;159:155-61. doi: 10.1111/j.1600-065x.1997.tb01013.x. Immunol Rev. 1997. PMID: 9416509 Review. - Selective autophagy and viruses.
Sumpter R Jr, Levine B. Sumpter R Jr, et al. Autophagy. 2011 Mar;7(3):260-5. doi: 10.4161/auto.7.3.14281. Autophagy. 2011. PMID: 21150267 Free PMC article. Review.
Cited by
- Species-specific impact of the autophagy machinery on Chikungunya virus infection.
Judith D, Mostowy S, Bourai M, Gangneux N, Lelek M, Lucas-Hourani M, Cayet N, Jacob Y, Prévost MC, Pierre P, Tangy F, Zimmer C, Vidalain PO, Couderc T, Lecuit M. Judith D, et al. EMBO Rep. 2013 Jun;14(6):534-44. doi: 10.1038/embor.2013.51. Epub 2013 Apr 26. EMBO Rep. 2013. PMID: 23619093 Free PMC article. - Atg16L1 deficiency confers protection from uropathogenic Escherichia coli infection in vivo.
Wang C, Mendonsa GR, Symington JW, Zhang Q, Cadwell K, Virgin HW, Mysorekar IU. Wang C, et al. Proc Natl Acad Sci U S A. 2012 Jul 3;109(27):11008-13. doi: 10.1073/pnas.1203952109. Epub 2012 Jun 19. Proc Natl Acad Sci U S A. 2012. PMID: 22715292 Free PMC article. - Rhinovirus and Innate Immune Function of Airway Epithelium.
Ganjian H, Rajput C, Elzoheiry M, Sajjan U. Ganjian H, et al. Front Cell Infect Microbiol. 2020 Jun 19;10:277. doi: 10.3389/fcimb.2020.00277. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32637363 Free PMC article. Review. - Macroautophagy--friend or foe of viral replication?
Münz C. Münz C. EMBO Rep. 2013 Jun;14(6):483-4. doi: 10.1038/embor.2013.55. Epub 2013 May 10. EMBO Rep. 2013. PMID: 23661081 Free PMC article. No abstract available. - Foot-and-mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3-kinase-independent pathway.
Berryman S, Brooks E, Burman A, Hawes P, Roberts R, Netherton C, Monaghan P, Whelband M, Cottam E, Elazar Z, Jackson T, Wileman T. Berryman S, et al. J Virol. 2012 Dec;86(23):12940-53. doi: 10.1128/JVI.00846-12. Epub 2012 Sep 19. J Virol. 2012. PMID: 22993157 Free PMC article.
References
- Gil-Fernandez C, Ronda-Lain C, Rubio-Huertos M. Electron microscopic study of Sindbis virus morphogenesis. Arch Gesamte Virusforsch. 1973;40:1–9. - PubMed
- Griffin DE. Neuronal cell death in alphavirus encephalomyelitis. In Current Topics in Microbiology and Immunology. 2005;289:57–77. - PubMed
- Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI151367/AI/NIAID NIH HHS/United States
- R01 AI051367-06/AI/NIAID NIH HHS/United States
- T32 AI007520/AI/NIAID NIH HHS/United States
- R01 AI151367/AI/NIAID NIH HHS/United States
- R01 AI051367/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources