Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies - PubMed (original) (raw)
Comparative Study
Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies
Jaime M Predmore et al. Circulation. 2010.
Abstract
Background: The ubiquitin proteasome system maintains a dynamic equilibrium of proteins and prevents accumulation of damaged and misfolded proteins, yet its role in human cardiac dysfunction is not well understood. The present study evaluated ubiquitin proteasome system function in human heart failure and hypertrophic cardiomyopathy (HCM).
Methods and results: Proteasome function was studied in human nonfailing donor hearts, explanted failing hearts, and myectomy samples from patients with HCM. Proteasome proteolytic activities were markedly reduced in failing and HCM hearts compared with nonfailing hearts (P<0.01). This activity was partially restored after mechanical unloading in failing hearts (P<0.01) and was significantly lower in HCM hearts with pathogenic sarcomere mutations than in those lacking these mutations (P<0.05). There were no changes in the protein content of ubiquitin proteasome system subunits (ie, 11S, 20S, and 19S) or in active-site labeling of the 20S proteolytic subunit beta-5 among groups to explain decreased ubiquitin proteasome system activity in HCM and failing hearts. Examination of protein oxidation revealed that total protein carbonyls, 4-hydroxynonenylated proteins, and oxidative modification to 19S ATPase subunit Rpt 5 were increased in failing compared with nonfailing hearts.
Conclusions: Proteasome activity in HCM and failing human hearts is impaired in the absence of changes in proteasome protein content or availability of proteolytic active sites. These data provide strong evidence that posttranslational modifications to the proteasome may account for defective protein degradation in human cardiomyopathies.
Conflict of interest statement
Disclosures: There are no conflicts to disclose
Figures
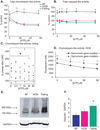
Figure 1. Cardiac 26S ubiquitin proteasome activity and ubiquinated protein accumulation in nonfailing, hypertrophic cardiomyopathy and end-stage failing hearts
NF=nonfailing, HCM=hypertrophic cardiomyopathy. All values are expressed as a mean ± SEM. A. Heart tissue homogenates (60 µg total cytosolic protein) were assayed for proteasome peptidase activity in the presence of ATP. Total chymotrypsin-like activity represents the difference in fluorescence in the presence and absence of the specific inhibitor, lactacystin (18 µmol/L). †P<0.05 for NF (n=6) vs HCM (n=17). *P<0.01 for NF vs HCM and NF vs Failing (n=14). **B**. Total caspase-like activity represents the difference in fluorescence in the presence and absence of the specific inhibitor, ZPNAC (10 µmol/L). *P<0.01 for NF (n=5) vs HCM (n=8) and NF vs Failing (n=8). **C**. Comparison of peak chymotrypsin-like activity in samples from the same patients (n=6) before and after implantation of a left ventricular assist device (LVAD). Each black square represents a single observation and red circle with line represents the mean % change in post-LVAD compared to pre-LVAD samples (*P<0.05). **D**. Comparison of chymotrypsin-like activity in samples from HCM patients with or without sarcomere gene mutations. *P<0.05 for mutation positive (n=6) vs mutation negative (n=7). **E**. Representative immunoblot of human total protein homogenates (50 µg) probed with an antibody against polyubiquinated proteins. **F**. Densitometric analysis for polyubiquinated proteins (>100KDa), standardized to GAPDH as a protein loading control. N=5 (NF), 11 (HCM), 10 (failing). *P<0.05 for NF vs failing.
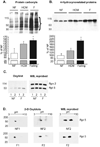
Figure 2. Akt and p53 expression in human whole heart homogenates
Representative immunoblots (left) and densitometric analysis (right) for Akt and p53. * P<0.05 for Akt and P<0.001 for p53 comparing both HCM and failing groups to non-failing.
Figure 3. Oxidative protein damage in normal, hypertrophic and failing human heart_s_
Samples were analyzed for (A) protein carbonyls after exposure to 2,4-dinitrophenylhydrazine (DNPH) and (B) 4-hydroxynonelated proteins. The depicted membranes are representative of 5 to 6 different samples in each group. Densitometric analysis is presented in the graphs beneath the membranes. In the protein carbonyl samples, overall average density was calculated over the range of 30 to 115 kDa. *P<0.05 for NF vs failing hearts. C. (Left) Proteasome subunit proteins from highly enriched 26S proteasome fractions were reacted with DNPH and probed with a DNPH-specific antibody. (Right) The membrane was stripped and reprobed with antibodies specific for Rpt5 and Rpn2. D. (Left and middle) 2D-gel electrophoresis of proteasome subunits previously reacted with DNPH. These membranes were probed with an antibody specific for DNPH. (Right) The membranes were stripped and reacted with an antibody to Rpt 5.
Similar articles
- Impaired assembly and post-translational regulation of 26S proteasome in human end-stage heart failure.
Day SM, Divald A, Wang P, Davis F, Bartolone S, Jones R, Powell SR. Day SM, et al. Circ Heart Fail. 2013 May;6(3):544-9. doi: 10.1161/CIRCHEARTFAILURE.112.000119. Epub 2013 Mar 20. Circ Heart Fail. 2013. PMID: 23515276 Free PMC article. - Energetics and function of the failing human heart with dilated or hypertrophic cardiomyopathy.
Kalsi KK, Smolenski RT, Pritchard RD, Khaghani A, Seymour AM, Yacoub MH. Kalsi KK, et al. Eur J Clin Invest. 1999 Jun;29(6):469-77. doi: 10.1046/j.1365-2362.1999.00468.x. Eur J Clin Invest. 1999. PMID: 10354207 - Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy.
Birks EJ, Latif N, Enesa K, Folkvang T, Luong le A, Sarathchandra P, Khan M, Ovaa H, Terracciano CM, Barton PJ, Yacoub MH, Evans PC. Birks EJ, et al. Cardiovasc Res. 2008 Aug 1;79(3):472-80. doi: 10.1093/cvr/cvn083. Epub 2008 Mar 28. Cardiovasc Res. 2008. PMID: 18375498 - Proteasome dysfunction in cardiomyopathies.
Gilda JE, Gomes AV. Gilda JE, et al. J Physiol. 2017 Jun 15;595(12):4051-4071. doi: 10.1113/JP273607. Epub 2017 Mar 16. J Physiol. 2017. PMID: 28181243 Free PMC article. Review. - The ubiquitin-proteasome system and nonsense-mediated mRNA decay in hypertrophic cardiomyopathy.
Carrier L, Schlossarek S, Willis MS, Eschenhagen T. Carrier L, et al. Cardiovasc Res. 2010 Jan 15;85(2):330-8. doi: 10.1093/cvr/cvp247. Epub 2009 Jul 17. Cardiovasc Res. 2010. PMID: 19617224 Free PMC article. Review.
Cited by
- The COP9 signalosome and cullin-RING ligases in the heart.
Wang X, Martin DS. Wang X, et al. Am J Cardiovasc Dis. 2015 Mar 20;5(1):1-18. eCollection 2015. Am J Cardiovasc Dis. 2015. PMID: 26064789 Free PMC article. Review. - Pregnancy is associated with decreased cardiac proteasome activity and oxidative stress in mice.
Iorga A, Dewey S, Partow-Navid R, Gomes AV, Eghbali M. Iorga A, et al. PLoS One. 2012;7(11):e48601. doi: 10.1371/journal.pone.0048601. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166589 Free PMC article. - Effects of MYBPC3 loss-of-function mutations preceding hypertrophic cardiomyopathy.
Helms AS, Tang VT, O'Leary TS, Friedline S, Wauchope M, Arora A, Wasserman AH, Smith ED, Lee LM, Wen XW, Shavit JA, Liu AP, Previs MJ, Day SM. Helms AS, et al. JCI Insight. 2020 Jan 30;5(2):e133782. doi: 10.1172/jci.insight.133782. JCI Insight. 2020. PMID: 31877118 Free PMC article. - Identification and functional analysis of senescent cells in the cardiovascular system using omics approaches.
Mahoney SA, Dey AK, Basisty N, Herman AB. Mahoney SA, et al. Am J Physiol Heart Circ Physiol. 2023 Nov 1;325(5):H1039-H1058. doi: 10.1152/ajpheart.00352.2023. Epub 2023 Sep 1. Am J Physiol Heart Circ Physiol. 2023. PMID: 37656130 Free PMC article. Review. - Rare variants in genes encoding MuRF1 and MuRF2 are modifiers of hypertrophic cardiomyopathy.
Su M, Wang J, Kang L, Wang Y, Zou Y, Feng X, Wang D, Ahmad F, Zhou X, Hui R, Song L. Su M, et al. Int J Mol Sci. 2014 May 26;15(6):9302-13. doi: 10.3390/ijms15069302. Int J Mol Sci. 2014. PMID: 24865491 Free PMC article.
References
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed
- Glickman MH, Raveh D. Proteasome plasticity. FEBS letters. 2005;579:3214–3223. - PubMed
- Zong C, Gomes AV, Drews O, Li X, Young GW, Berhane B, Qiao X, French SW, Bardag-Gorce F, Ping P. Regulation of murine cardiac 20S proteasomes: role of associating partners. Circ Res. 2006;99:372–380. - PubMed
- Mason GG, Hendil KB, Rivett AJ. Phosphorylation of proteasomes in mammalian cells. Identification of two phosphorylated subunits and the effect of phosphorylation on activity. Eur J Biochem. 1996;238:453–462. - PubMed
- Satoh K, Sasajima H, Nyoumura KI, Yokosawa H, Sawada H. Assembly of the 26S proteasome is regulated by phosphorylation of the p45/Rpt6 ATPase subunit. Biochemistry. 2001;40:314–319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources