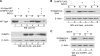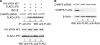The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process - PubMed (original) (raw)
The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process
Andrew Pincetic et al. J Virol. 2010 May.
Abstract
The release of retroviruses from cells requires ubiquitination of Gag and recruitment of cellular proteins involved in endosome sorting, including the ESCRT-III proteins and the Vps4 ATPase. In response to infection, cells have evolved an interferon-induced mechanism to block virus replication through expression of the interferon-stimulated gene 15 (ISG15), a dimer homologue of ubiquitin, which interferes with ubiquitin pathways in cells. Previously, it has been reported that ISG15 expression inhibited the E3 ubiquitin ligase, Nedd4, and prevented association of the ESCRT-I protein Tsg101 with human immunodeficiency virus type 1 (HIV-1) Gag. The budding of avian sarcoma leukosis virus and HIV-1 Gag virus-like particles containing L-domain mutations can be rescued by fusion to ESCRT proteins, which cause entry into the budding pathway beyond these early steps. The release of these fusions from cells was susceptible to inhibition by ISG15, indicating that there was a block late in the budding process. We now demonstrate that the Vps4 protein does not associate with the avian sarcoma leukosis virus or the HIV-1 budding complexes when ISG15 is expressed. This is caused by a loss in interaction between Vps4 with its coactivator protein LIP5 needed to promote the formation of the ESCRT-III-Vps4 double-hexamer complex required for membrane scission and virus release. The inability of LIP5 to interact with Vps4 is the probable result of ISG15 conjugation to the ESCRT-III protein, CHMP5, which regulates the availability of LIP5. Thus, there appear to be multiple levels of ISG15-induced inhibition acting at different stages of the virus release process.
Figures
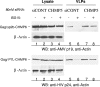
FIG. 1.
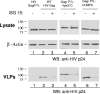
Expression of ISG15 inhibited ASLV VLP release from cells. (A) 293/E cells were cotransfected with p2036 plasmids (0.5 μg of each) expressing wild-type ASLV Gag, UBE1L, and UBCH8 (lanes 1 to 6) with increasing concentrations of pISG15 (indicated as μg of plasmid DNA transfected into cells. At 48 h posttransfection, Gag was immunoprecipitated from cell extracts. VLPs were collected from the medium fraction by pelleting through a 20% sucrose cushion as described in Materials and Methods. Samples were fractionated by SDS-10% PAGE and immunoblotted with anti-AMV MA (p19) or anti-ISG15 sera. Anti-actin serum was used to detect β-actin as a loading control. (B). 293/E cells were transfected with the p2036 plasmid expressing wild-type Gag (0.5 μg) and the pISG15 plasmid expressing ISG15 (3 μg). Samples were analyzed as in panel A.
FIG. 2.
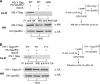
ISG15 does not block ASLV Gag-Nedd4 association. 293/E cells were transfected with p2036 expressing wild-type ASLV Gag (lanes 1 and 2) or ASLV Gag/Δp2b (lane 3) and, where indicated, with 3 μg of pISG15 (lane 2). At 48 h posttransfection, lysate fractions were immunoprecipitated with anti-AMV MA (p19) serum, and VLPs were collected from the medium fraction as described in the legend to Fig. 1. Samples were resolved by SDS-10% PAGE. Endogenous Nedd4 precipitating with Gag was detected by Western blotting with an anti-Nedd4 serum. Gag in the lysate fraction and VLPs were detected by Western blotting with anti-AMV MA (p19) serum. ISG15 was detected as in legend to Fig. 1.
FIG. 3.
ISG15 inhibited budding of HIV-1 Gag-GFP and HIV-1 Gag/P7L-ESCRT chimeras. 293/E cells were transfected with p2036 expressing HIV-1 Gag/P7L-GFP (lane 1), Gag-GFP (lanes 2 and 3), Gag/P7L-Vps37C (lanes 4 and 5), or Gag/P7L-CHMP6 (lanes 6 and 7) and, where indicated, with 3 μg of pISG15 (lanes 3, 5, and 7). At 48 h posttransfection, Gag was collected as described in the legend to Fig. 1. Gag expression in lysates and VLP release into the media was detected by Western blotting with anti-HIV-1 CA (p24) serum. All other notations are as in legend to Fig. 1. The positions of migration of molecular weight markers are indicated on the left side of the figures.
FIG. 4.
ISG15 inhibited budding of wild-type ASLV Gag and ASLV Gag/Δp2b-ESCRT chimeras. 293/E cells were transfected with p2036 expressing ASLV Gag/Δp2b (lane 1), wild-type ASLV Gag (lanes 2 and 3), Gag/Δp2b-Vps37C (lanes 4 and 5), Gag/Δp2b-Eap20 (lanes 6 and 7), or Gag/Δp2b-CHMP6 (lanes 8 and 9) and, where indicated, 3 μg of pISG15. At 48 h posttransfection, Gag was collected from lysate and medium fractions and analyzed as described in the legend to Fig. 1. Gag was detected by Western blotting with an anti-MA (p19) serum.
FIG. 5.
ASLV and HIV-1 Gag failed to associate with Vps4EQ in the presence of ISG15. (A) 293/E cells were cotransfected with p2036 expressing wild-type ASLV Gag (lanes 1 and 2) or Gag/Δp2b (lane 3) and HA-Vps4E228Q (lanes 1 to 3) and, where indicated, pISG15 (lane 2). At 48 h posttransfection, 10% of total cell lysate input was resolved by SDS-10% PAGE to verify the expression of Gag and HA-Vps4EQ by Western blotting with anti-HA or AMV MA serum (bottom panel, marked WB). HA-Vps4EQ was immunoprecipitated with anti-HA antibody from remaining cell lysate fraction and resolved by SDS-10% PAGE. Gag was detected by Western blotting with an antiserum directed at the AMV MA protein (top panel, marked Co-IP). HA-Vps4EQ was detected with the anti-HA serum. (B) 293/E cells were cotransfected with p2036 expressing HIV-1 Gag-GFP (lanes 1 and 2) or HIV-1 Gag/P7L-GFP (lane 3) and HA-Vps4E228Q (lanes 1 to 3). An expression plasmid for pISG15 was cotransfected into cells where indicated (lane 2). At 48 h posttransfection, 10% of the total cell lysate input was resolved by SDS-10% PAGE to verify the expression of Gag and HA-Vps4EQ (bottom panel, marked WB). HA-Vps4EQ was immunoprecipitated with anti-HA serum from remaining cell lysate fraction and resolved by SDS-10% PAGE. Gag was detected by Western blotting with anti-HIV-1 CA (p24) serum (top panel marked, Co-IP). HA-Vps4EQ was detected with anti-HA serum.
FIG. 6.
ISG15 conjugated to the ESCRT-III protein, CHMP5. 293/E cells were transfected with or without plasmids expressing His6-HA-ISG15, UbE1L, and UbsH8 as indicated. At 48 h posttransfection, cell extracts were resolved by SDS-10% PAGE. (Top panel) Proteins were detected with an anti-CHMP5 serum by Western blotting. The lower and upper bands observed correspond in size to the endogenous CHMP5 and its ISG15 conjugate, respectively. (Middle panel) Total cell lysate (10% of input) was resolved by SDS-10% PAGE to verify expression of His6-HA-ISG15. (Bottom panel) Blot from panel A was washed and reprobed with an anti-HA serum to detect the His6-HA-ISG15-conjugated CHMP5.
FIG. 7.
ISG15 altered membrane association of Vps4 and CHMP5. (A) 293/E cells were transfected with p2036 encoding WT HA-Vps4 and, where indicated, with plasmids expressing CHMP5-FLAG and ISG15. At 48 h posttransfection, cells were lysed, and soluble and pelleted fractions were prepared from the lysate fraction as described in Materials and Methods. Distribution of CHMP5-FLAG and HA-Vps4 WT in the resulting soluble (S) (lanes 1, 3, and 5) and the membrane-bound pellet (P) (lanes 2, 4, and 6) fractions were visualized by Western blotting. (Top panel) Blot probed with anti-HA serum to detect HA-Vps4. (Bottom panel) Blot probed with anti-FLAG serum to detect CHMP5-FLAG expression. β-Actin served as a loading control. (B) 293E cells were transfected with plasmid expressing CHMP5-FLAG and ISG15 where indicated. Cells were lysed and soluble and pellet fractions were prepared as described in panel A. Endogenous Vps4 was detected with an antiserum directed at Vps4. (C) 293E cells were transfected with a plasmid expressing ISG15 where indicated, and samples were prepared as described in panel A. Endogenous CHMP5 was detected with an anti-CHMP5 serum.
FIG. 8.
ISG15 interfered with LIP5-Vps4 interaction. 293/E cells were cotransfected with plasmids expressing HA-Vps4 and FLAG-tagged LIP5 and increasing concentrations of pISG15 where indicated. At 48 h posttransfection, HA-Vps4 was immunoprecipitated from cell extracts with a mouse anti-HA serum, and samples were resolved by SDS-10% PAGE. For the top panel, precipitation of FLAG-LIP5 was determined by Western blotting with anti-FLAG antibody and goat anti-mouse IgG-HRP secondary antibody. The bands above and below FLAG-LIP5 are IgG heavy and light chains, respectively. The bottom panel shows a Western blot of the 10% input of the total cell lysates used in the coimmunoprecipitation assay.
FIG. 9.
CHMP5 is required for ISG15-mediated interference of LIP5-Vps4 interaction. (A) 293/E cells were cotransfected with plasmids encoding HA-Vps4, FLAG-LIP5, and ISG15, along with 40 (lane 2) or 80 (lane 3) nM concentrations of a siRNA pool directed against CHMP5 or 80 nM (lanes 1 and 4) of a nontargeting siRNA (siCONT). HA-tagged Vps4 proteins were immunoprecipitated from cell lysate fractions with an anti-HA serum, and precipitated proteins were resolved by SDS-10% PAGE. FLAG-LIP5 was detected by Western blotting with an anti-FLAG antibody. Expression of HA-Vps4 and FLAG-LIP5 were detected as described in the legend to Fig. 8 using anti-HA and FLAG serum. (B) CHMP5-FLAG was expressed in cells in the presence of siCONT (80 nM, lane 1) or the siRNA targeting CHMP5 (20 nM, lane 2; 40 nM, lane 3; 80 nM, lane 4), and proteins were analyzed as in panel A using an anti-FLAG serum. β-Actin again served as a loading control.
FIG. 10.
CHMP5 is required for ISG15-mediated interference of budding by ASLV Gag/Δp2b-CHMP6 or HIV-1 Gag/P7L-CHMP6. 293/E cells were cotransfected with plasmids encoding ASLV Gag/Δp2b-CHMP6 (top panel) or HIV-1 Gag/P7L-CHMP6 (bottom panel) with the ISG15 expression plasmid and control or CHMP5-specific siRNAs (80 nM) as indicated. Lysate and medium fractions were prepared and analyzed as described in legends to Fig. 3 and 4. β-Actin served as a gel loading control.
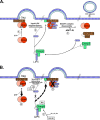
FIG. 11.
Model of the role of Vps4 in retrovirus budding. (A) Vps4 activity is regulated by the oligomeric state of the enzyme. In its inactive form, Vps4 exists as a dimer in the cytosol. (Step 1) Upon polymerization of the ESCRT-III complex with Gag, the ATP-bound Vps4 dimer is recruited to membranes through its interaction with MIM domains found in most ESCRT-III proteins. (Step 2) ATP-bound Vps4 dimers coassemble into double hexameric ring complexes with the coactivator protein, LIP5, to form the active enzyme. LIP5 binds the ESCRT-III protein, CHMP5, in the cytosol. Although CHMP5 can localize to membranes, the literature is unclear whether the LIP5-CHMP5 interaction persists once LIP5 coassembles with Vps4. (Step 3) ATP hydrolysis by the Vps4-LIP5 oligomer releases the ESCRT complexes from membranes and returns Vps4 to its inactive dimeric form in the cytoplasm. (Step 4) Dissociation of the ESCRT complex coincides with the membrane fission event to release retrovirus particles from the cell surface. (B) ISG15 prevents Vps4-LIP5 oligomerization. (Step1) The ESCRT-III protein, CHMP5, binds to LIP5 in the cytosol possibly preventing it from binding to MIM domains of the ESCRT-III complex. In the presence of ISG15-specific E1 and E2 enzymes, ISG15 conjugates to CHMP5. (Step 2) ISG15 overexpression blocks the interaction between Vps4 and its coactivator, LIP5. Although the function of CHMP5 remains largely uncharacterized, ISGylation of CHMP5 may alter Vps4-LIP5 complex assembly by sequestering LIP5. (Step 3) In the absence of the Vps4-LIP5 oligomer, ESCRT complexes remain bound to membranes and virus budding is arrested. (Step 4) Vps4 is released into the cytosol.
Similar articles
- Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15.
Kuang Z, Seo EJ, Leis J. Kuang Z, et al. J Virol. 2011 Jul;85(14):7153-61. doi: 10.1128/JVI.02610-10. Epub 2011 May 4. J Virol. 2011. PMID: 21543490 Free PMC article. - The functionally exchangeable L domains in RSV and HIV-1 Gag direct particle release through pathways linked by Tsg101.
Medina G, Zhang Y, Tang Y, Gottwein E, Vana ML, Bouamr F, Leis J, Carter CA. Medina G, et al. Traffic. 2005 Oct;6(10):880-94. doi: 10.1111/j.1600-0854.2005.00323.x. Traffic. 2005. PMID: 16138902 Free PMC article. - Interactions of the human LIP5 regulatory protein with endosomal sorting complexes required for transport.
Skalicky JJ, Arii J, Wenzel DM, Stubblefield WM, Katsuyama A, Uter NT, Bajorek M, Myszka DG, Sundquist WI. Skalicky JJ, et al. J Biol Chem. 2012 Dec 21;287(52):43910-26. doi: 10.1074/jbc.M112.417899. Epub 2012 Oct 26. J Biol Chem. 2012. PMID: 23105106 Free PMC article. - The regulation of Endosomal Sorting Complex Required for Transport and accessory proteins in multivesicular body sorting and enveloped viral budding - An overview.
Ahmed I, Akram Z, Iqbal HMN, Munn AL. Ahmed I, et al. Int J Biol Macromol. 2019 Apr 15;127:1-11. doi: 10.1016/j.ijbiomac.2019.01.015. Epub 2019 Jan 4. Int J Biol Macromol. 2019. PMID: 30615963 Review. - The antiviral activities of ISG15.
Morales DJ, Lenschow DJ. Morales DJ, et al. J Mol Biol. 2013 Dec 13;425(24):4995-5008. doi: 10.1016/j.jmb.2013.09.041. Epub 2013 Oct 3. J Mol Biol. 2013. PMID: 24095857 Free PMC article. Review.
Cited by
- New and novel intrinsic host repressive factors against HIV-1: PAF1 complex, HERC5 and others.
Tyagi M, Kashanchi F. Tyagi M, et al. Retrovirology. 2012 Mar 9;9:19. doi: 10.1186/1742-4690-9-19. Retrovirology. 2012. PMID: 22405323 Free PMC article. No abstract available. - ISG15 deficiency and increased viral resistance in humans but not mice.
Speer SD, Li Z, Buta S, Payelle-Brogard B, Qian L, Vigant F, Rubino E, Gardner TJ, Wedeking T, Hermann M, Duehr J, Sanal O, Tezcan I, Mansouri N, Tabarsi P, Mansouri D, Francois-Newton V, Daussy CF, Rodriguez MR, Lenschow DJ, Freiberg AN, Tortorella D, Piehler J, Lee B, García-Sastre A, Pellegrini S, Bogunovic D. Speer SD, et al. Nat Commun. 2016 May 19;7:11496. doi: 10.1038/ncomms11496. Nat Commun. 2016. PMID: 27193971 Free PMC article. - Host factors involved in retroviral budding and release.
Martin-Serrano J, Neil SJ. Martin-Serrano J, et al. Nat Rev Microbiol. 2011 Jun 16;9(7):519-31. doi: 10.1038/nrmicro2596. Nat Rev Microbiol. 2011. PMID: 21677686 Review. - Synthetic Lethal Interaction between the ESCRT Paralog Enzymes VPS4A and VPS4B in Cancers Harboring Loss of Chromosome 18q or 16q.
Neggers JE, Paolella BR, Asfaw A, Rothberg MV, Skipper TA, Yang A, Kalekar RL, Krill-Burger JM, Dharia NV, Kugener G, Kalfon J, Yuan C, Dumont N, Gonzalez A, Abdusamad M, Li YY, Spurr LF, Wu WW, Durbin AD, Wolpin BM, Piccioni F, Root DE, Boehm JS, Cherniack AD, Tsherniak A, Hong AL, Hahn WC, Stegmaier K, Golub TR, Vazquez F, Aguirre AJ. Neggers JE, et al. Cell Rep. 2020 Dec 15;33(11):108493. doi: 10.1016/j.celrep.2020.108493. Cell Rep. 2020. PMID: 33326793 Free PMC article. - Ilaprazole and other novel prazole-based compounds that bind Tsg101 inhibit viral budding of HSV-1/2 and HIV from cells.
Leis J, Luan CH, Audia JE, Dunne SF, Heath CM. Leis J, et al. J Virol. 2021 May 10;95(11):e00190-21. doi: 10.1128/JVI.00190-21. Epub 2021 Mar 17. J Virol. 2021. PMID: 33731460 Free PMC article.
References
- Arguello, M. D., and J. Hiscott. 2007. Ub surprised: viral ovarian tumor domain proteases remove ubiquitin and ISG15 conjugates. Cell Host Microbe 2:367-369. - PubMed
- Azmi, I. F., B. A. Davies, J. Xiao, M. Babst, Z. Xu, and D. J. Katzmann. 2008. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell 14:50-61. - PubMed
- Chang, Y.-G., X.-Z. Yan, Y.-Y. Xie, X.-C. Gao, A.-X. Song, D.-E. Zhang, and H.-Y. Hu. 2008. Different roles for two ubiquitin-like domains of ISG15 in protein modification. J. Biol. Chem. 283:13370-13377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous