Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions - PubMed (original) (raw)
Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions
David R Raleigh et al. Mol Biol Cell. 2010.
Abstract
In vitro studies have demonstrated that occludin and tricellulin are important for tight junction barrier function, but in vivo data suggest that loss of these proteins can be overcome. The presence of a heretofore unknown, yet related, protein could explain these observations. Here, we report marvelD3, a novel tight junction protein that, like occludin and tricellulin, contains a conserved four-transmembrane MARVEL (MAL and related proteins for vesicle trafficking and membrane link) domain. Phylogenetic tree reconstruction; analysis of RNA and protein tissue distribution; immunofluorescent and electron microscopic examination of subcellular localization; characterization of intracellular trafficking, protein interactions, dynamic behavior, and siRNA knockdown effects; and description of remodeling after in vivo immune activation show that marvelD3, occludin, and tricellulin have distinct but overlapping functions at the tight junction. Although marvelD3 is able to partially compensate for occludin or tricellulin loss, it cannot fully restore function. We conclude that marvelD3, occludin, and tricellulin define the tight junction-associated MARVEL protein family. The data further suggest that these proteins are best considered as a group with both redundant and unique contributions to epithelial function and tight junction regulation.
Figures
Figure 1.
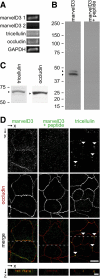
MarvelD3, tricellulin, and occludin are the only members of the tight junction–associated MARVEL protein (TAMP) subfamily. (A) MARVEL domain alignment was ordered after model fitting and tree reconstruction and shaded within each of the four MARVEL protein lineages according to the Blosum62 score. Amino acids are shaded according to physiochemical properties as indicated in the figure. The characteristic acidic residue in the third transmembrane domain (Sanchez-Pulido et al., 2002) is indicated by an asterisk (*). (B) The alignment was used for tree reconstruction, and disparities were calculated according to the fraction of mutated residues. Posterior values are shown as 100-fold original predictions, implying that a bootstrap value of 60 is highly reliable. (C) Gene conservation analysis demonstrates that exons encoding MARVEL domains (∗) show the highest degree of evolutionary conservation within the marvelD3, tricellulin, and occludin genes. Data are presented as a base-by-base conservation measurement, with 0 indicating a neutral region and 1 indicating perfect conservation, from 28 vertebrate species including marsupial, monotreme, and placental mammals, as well as reptile, amphibian, bird, and fish clades. (D) Analysis of transcript expression in the Affymetrix GeneChip Mouse Genome 430 2.0 Array (Santa Clara, CA) reveals a highly correlated (p < 0.001), though nonredundant, pattern of expression for marvelD3, tricellulin, and occludin within epithelial organs. (E) EGFP-MARVEL domain–containing fusion proteins (green) were transfected into Caco-2 human intestinal epithelial cells. Polarized monolayers were grown on Transwell supports, and ZO-1 (red) was detected by immunofluorescent staining. Three-dimensional reconstruction shows that EGFP-marvelD3 splice variants 1 and 2, EGFP-tricellulin, and EGFP-occludin localize to tight junctions and intracellular vesicles. Both EGFP-marvelD3 splice variants are enriched along bicellular tight junctions, similar to occludin, whereas tricellulin is specifically concentrated at tricellular regions. MARVEL proteins from other lineages were found to decorate the apical plasma membrane and intracellular transport vesicles, but not the tight junction. Images are representative of ≥3 experiments, all with similar results. Bar, 10 μm.
Figure 2.
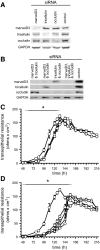
TAMPs are differentially expressed at intercellular junctions of Caco-2 human intestinal epithelial cell monolayers. (A) Analysis of TAMP transcript expression by RT-PCR reveals differential expression of marvelD3 splice variants, tricellulin, and occludin in Caco-2 cells. (B) MarvelD3 proteins (arrowheads) were detected in Caco-2 cells as determined by SDS-PAGE immunoblot analysis. Preincubation of polyclonal antisera with peptides used as immunogens eliminated the marvelD3 bands. (C) Tricellulin and occludin proteins (arrowheads) were also detected in Caco-2 cells by SDS-PAGE immunoblot. (D) Immunofluorescence of polarized Caco-2 monolayers demonstrates junctional and vesicular localization of all TAMP family members. MarvelD3 (green) staining was abrogated following preincubation of antisera with competing peptides. Tricellulin (green) specifically localized to tricellular junctions (arrowheads). Occludin (red) is shown for orientation in each image. Bar,10 μm. Imaging along the _z_-axis at the indicated position (dashed line) reveals colocalization of marvelD3 and tricellulin with occludin at apical intercellular junctions. Bar, 5 μm.
Figure 3.
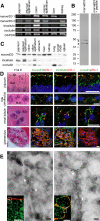
TAMPs are differentially expressed across epithelial tissues. (A) Analysis of TAMP transcript expression by semiquantitative RT-PCR reveals differential expression of marvelD3 splice variants, tricellulin, and occludin message in epithelial-rich mouse organs. (B) MarvelD3 splice variants (arrowheads) were each detected in mouse jejunal epithelium samples as determined by SDS-PAGE immunoblot analysis. Preincubation of polyclonal antisera with peptides used as immunogens eliminated the marvelD3 bands. (C) Analysis of TAMP expression across an array of mouse organs showed that marvelD3, tricellulin, and occludin proteins are differentially expressed in epithelium-rich tissues. (D) Immunofluorescence shows differential localization of marvelD3, tricellulin, and occludin (green) in vivo. All TAMP family members are concentrated at the apical intercellular junction of jejunal epithelia, hepatocytes, and renal tubular epithelium. In contrast, glomerular epithelial cells are enriched in marvelD3 and tricellulin, but largely lack occludin immunofluorescence. Nuclei (blue), ZO-1 (red), and H&E images (BB, brush border; TJ, tight junction; L, lumen; BLM, basolateral membrane; N, nucleus; BC, Bowman's capsule; M, mesenchyme; GT, glomerular tuft; US, urinary space) are shown for orientation. Bars, 10 μm. (E) Transmission immunoelectron microscopy of mouse jejunum demonstrates specific localization of marvelD3 to the tight junction in sagittal and en face orientations (arrowheads). Immunofluorescence insets with marvelD3 (green) and actin (red) are shown for orientation. Gray boxes approximate electron microscopic fields. White bars, 10 μm; black bars, 100 nm. Data are representative of ≥2 experiments, all with similar results.
Figure 4.
TAMPs demonstrate unique interactions with each other and ZO-1. (A) Coimmunoprecipitation of TAMPs from Caco-2 lysates reveals interactions among marvelD3, tricellulin, and occludin. The data are consistent with a model of TAMP interaction, where marvelD3 physically associates with both occludin and tricellulin, but the latter two TAMPs do not interact. (B) TAMP cytoplasmic tails were expressed as GST fusion constructs, and equal quantities of each were loaded onto GST-agarose, as confirmed by the Coomassie blue–stained gel. Pulldown experiments using VSVG-ZO-1-GuK as prey demonstrate interaction with the carboxy-terminal tail of occludin (immunoblot). In contrast, neither cytoplasmic tails of tricellulin nor marvelD3 were able to pull down VSVG-ZO-1-GuK. Data are representative of ≥2 experiments, each in duplicate, with similar results.
Figure 5.
TAMPs partition into detergent-insoluble membrane microdomains and claudin-based tight junction-like strands. (A) Caco-2 monolayers were solubilized at 4°C in Triton X-100, and lysates were separated on sucrose density gradients. OD600, a measurement of the light scattering property of intact membranes, and protein recovery, are shown as a function of density. (B) Immunoblot analysis demonstrated that TAMPs are enriched in the light-scattering fraction of detergent-resistant membranes. (C) EGFP-TAMP constructs (green) were transfected into L929 human fibroblast cells alone or with mRFP1-claudin-1 (red). Monolayers were grown on glass coverslips, and nuclei (blue) were detected by staining with Hoechst 33342. EGFP-marvelD3 splice variants 1 and 2, EGFP-tricellulin, and EGFP-occludin localize to intracellular vesicles when expressed independently, but are recruited to tight junction-like strands (yellow) along areas of membrane overlap (arrowheads) in cells expressing claudin-1. Images are representative of ≥3 experiments, all with similar results. Bar, 10 μm.
Figure 6.
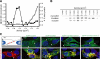
Epithelial barrier development coincides with TAMP expression and trafficking to the tight junction. (A) TER was monitored during barrier development in Caco-2 monolayers. Cells were collected for qRT-PCR, immunofluorescence, and SDS-PAGE immunoblot analyses at the indicated times. (B) qRT-PCR data show that tricellulin and marvelD3 splice variant 1 mRNA synthesis is induced during TER development. (C) SDS-PAGE immunoblot shows that TAMP protein expression increases during barrier development. (D) Immunofluorescence of Caco-2 monolayers demonstrates that marvelD3, tricellulin, occludin, and claudin-1 traffic to the tight junction during TER development. Matched exposures are shown for each protein. Arrowheads indicate tricellular regions. Bar, 20 μm. Data are representative of ≥3 experiments, each in duplicate, all with similar results.
Figure 7.
TAMP knockdown delays tight junction assembly. Caco-2 cells were treated with either control or TAMP-targeted siRNAs and plated on Transwell supports. (A and B) SDS-PAGE immunoblot analysis revealed that TAMP-targeted siRNAs specifically knocked down expression of only the targeted TAMP and that treatment was not associated with compensatory upregulation of other family members. (C) Knockdown (KD) of single TAMPs (marvelD3 KD, ▿; tricellulin KD, □; occludin KD, ◇) significantly (*p < 0.01) delayed barrier development relative to control monolayers (○). (D) Simultaneous knockdown of multiple TAMPs (control, ○; marvelD3 and tricellulin KD, ▿; marvelD3 and occludin KD, □; tricellulin and occludin KD, ◇; marvelD3, tricellulin and occludin KD, ▵) produced greater delays than loss of any one TAMP (*p < 0.05). Solid line: significant TER delay for all knockdown conditions; dashed line: significant TER delay for at least one knockdown condition. Data are representative of ≥5 experiments, each in quadruplicate, all with similar results.
Figure 8.
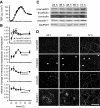
TAMPs have unique dynamic behaviors at bicellular and tricellular tight junction regions. (A) Caco-2 cells were transfected with EGFP-TAMP fusion proteins, and protein dynamics at bicellular (bTJ) and tricellular (tTJ) tight junction regions were assessed by FRAP. TAMPs exhibit distinct and unique dynamic behaviors at tricellular (B–D) and bicellular (E–G) areas of the tight junction. Occludin and tricellulin are highly mobile at both regions. In contrast, mobility of both marvelD3 splice variants varied between bicellular and tricellular tight junction regions (see Table 1). Data are representative of ≥3 experiments, each in triplicate, all with similar results.
Figure 9.
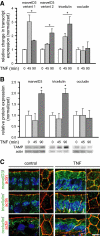
TAMPs are differentially expressed and redistributed in response to in vivo TNF treatment. Mice were sacrificed at the indicated times after intraperitoneal injection of 5 μg of TNF. (A) Epithelial cells were isolated, and RNA was harvested for qRT-PCR analysis. MarvelD3 and tricellulin message increased 45 min after TNF treatment and remained elevated after 90 min (*p < 0.01), whereas occludin message content was not changed (p > 0.40). (B) SDS-PAGE immunoblot and densitometric analyses of jejunal epithelial lysates demonstrate increased marvelD3 and tricellulin protein after 90 min of TNF exposure (*p < 0.05). (C) Jejunum was frozen 120 min after TNF injection and immunostained for TAMPs (green), F-actin (red), and nuclei (blue). MarvelD3 and tricellulin protein expression increased and was enriched at the tight junction, lateral membrane, and cytoplasm. Bar, 5 μm. Data are representative of ≥3 experiments, all with similar results.
Figure 10.
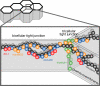
Model of TAMP locations and interactions at the tight junction. The figure shows the organization of proteins within a small area of a confluent monolayer (see image at the top). The bicellular tight junction is the interface between two cells, whereas the vertex where three cells meet is termed the tricellular tight junction. The tight junction strands within both bicellular and tricellular regions are composed of claudins (black spheres). All three TAMP proteins, marvelD3 (orange spheres), tricellulin (green spheres), and occludin (blue spheres) incorporate into claudin-based tight junction strands. Occludin and tricellulin are primarily found at bicellular and tricellular regions, respectively, whereas marvelD3 is present at both sites. This may explain why marvelD3 binds to both tricellulin and occludin, but tricellulin and occludin do not interact with one another. Tricellulin is unique in that it is present at the tight junction and along the lateral membrane. Only occludin interacts with the ZO-1 (red spheres) via the GuK domain. Thus, although there are similarities, each TAMP can also be defined by unique characteristics.
Similar articles
- Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family.
Steed E, Rodrigues NT, Balda MS, Matter K. Steed E, et al. BMC Cell Biol. 2009 Dec 22;10:95. doi: 10.1186/1471-2121-10-95. BMC Cell Biol. 2009. PMID: 20028514 Free PMC article. - In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization.
Cording J, Berg J, Käding N, Bellmann C, Tscheik C, Westphal JK, Milatz S, Günzel D, Wolburg H, Piontek J, Huber O, Blasig IE. Cording J, et al. J Cell Sci. 2013 Jan 15;126(Pt 2):554-64. doi: 10.1242/jcs.114306. Epub 2012 Nov 30. J Cell Sci. 2013. PMID: 23203797 - Downregulation of tight junction-associated MARVEL protein marvelD3 during epithelial-mesenchymal transition in human pancreatic cancer cells.
Kojima T, Takasawa A, Kyuno D, Ito T, Yamaguchi H, Hirata K, Tsujiwaki M, Murata M, Tanaka S, Sawada N. Kojima T, et al. Exp Cell Res. 2011 Oct 1;317(16):2288-98. doi: 10.1016/j.yexcr.2011.06.020. Epub 2011 Jul 8. Exp Cell Res. 2011. PMID: 21763689 - A look at tricellulin and its role in tight junction formation and maintenance.
Mariano C, Sasaki H, Brites D, Brito MA. Mariano C, et al. Eur J Cell Biol. 2011 Oct;90(10):787-96. doi: 10.1016/j.ejcb.2011.06.005. Epub 2011 Aug 24. Eur J Cell Biol. 2011. PMID: 21868126 Review. - Modulation of tight junction structure and function by kinases and phosphatases targeting occludin.
Dörfel MJ, Huber O. Dörfel MJ, et al. J Biomed Biotechnol. 2012;2012:807356. doi: 10.1155/2012/807356. Epub 2012 Jan 23. J Biomed Biotechnol. 2012. PMID: 22315516 Free PMC article. Review.
Cited by
- Tight junction proteins: from barrier to tumorigenesis.
Runkle EA, Mu D. Runkle EA, et al. Cancer Lett. 2013 Aug 28;337(1):41-8. doi: 10.1016/j.canlet.2013.05.038. Epub 2013 Jun 3. Cancer Lett. 2013. PMID: 23743355 Free PMC article. Review. - Viral interactions with the blood-brain barrier: old dog, new tricks.
Hou J, Baker LA, Zhou L, Klein RS. Hou J, et al. Tissue Barriers. 2016 Jan 28;4(1):e1142492. doi: 10.1080/21688370.2016.1142492. eCollection 2016 Jan-Mar. Tissue Barriers. 2016. PMID: 27141421 Free PMC article. Review. - Up-Regulation of Claudin-6 in the Distal Lung Impacts Secondhand Smoke-Induced Inflammation.
Lewis JB, Milner DC, Lewis AL, Dunaway TM, Egbert KM, Albright SC, Merrell BJ, Monson TD, Broberg DS, Gassman JR, Thomas DB, Arroyo JA, Reynolds PR. Lewis JB, et al. Int J Environ Res Public Health. 2016 Oct 17;13(10):1018. doi: 10.3390/ijerph13101018. Int J Environ Res Public Health. 2016. PMID: 27763528 Free PMC article. - Dynamic migration of γδ intraepithelial lymphocytes requires occludin.
Edelblum KL, Shen L, Weber CR, Marchiando AM, Clay BS, Wang Y, Prinz I, Malissen B, Sperling AI, Turner JR. Edelblum KL, et al. Proc Natl Acad Sci U S A. 2012 May 1;109(18):7097-102. doi: 10.1073/pnas.1112519109. Epub 2012 Apr 17. Proc Natl Acad Sci U S A. 2012. PMID: 22511722 Free PMC article. - Tight junction-related human diseases.
Sawada N. Sawada N. Pathol Int. 2013 Jan;63(1):1-12. doi: 10.1111/pin.12021. Epub 2013 Jan 7. Pathol Int. 2013. PMID: 23356220 Free PMC article. Review.
References
- Abascal F., Zardoya R., Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
- Abramoff M. D., Magelhaes P. J., Ram S. J. Image processing with Image. J. Biophoton. Int. 2004;11:36–42.
- Balda M. S., Whitney J. A., Flores C., Gonzalez S., Cereijido M., Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol. 1996;134:1031–1049. - PMC - PubMed
- Breitwieser G. E., McLenithan J. C., Cortese J. F., Shields J. M., Oliva M. M., Majewski J. L., Machamer C. E., Yang V. W. Colonic epithelium-enriched protein A4 is a proteolipid that exhibits ion channel characteristics. Am. J. Physiol. 1997;272:C957–C965. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 DK067887/DK/NIDDK NIH HHS/United States
- P30CA14599/CA/NCI NIH HHS/United States
- P30 CA014599/CA/NCI NIH HHS/United States
- T32 HL007237/HL/NHLBI NIH HHS/United States
- R01 DK061931/DK/NIDDK NIH HHS/United States
- R01 DK068271/DK/NIDDK NIH HHS/United States
- R01DK61931/DK/NIDDK NIH HHS/United States
- T32HL007237/HL/NHLBI NIH HHS/United States
- P01DK67887/DK/NIDDK NIH HHS/United States
- R01DK68271/DK/NIDDK NIH HHS/United States
- T32GM07281/GM/NIGMS NIH HHS/United States
- T32 GM007281/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous