Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79 - PubMed (original) (raw)
Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79
Atteeq Ur Rehman et al. Am J Hum Genet. 2010.
Abstract
Targeted genome capture combined with next-generation sequencing was used to analyze 2.9 Mb of the DFNB79 interval on chromosome 9q34.3, which includes 108 candidate genes. Genomic DNA from an affected member of a consanguineous family segregating recessive, nonsyndromic hearing loss was used to make a library of fragments covering the DFNB79 linkage interval defined by genetic analyses of four pedigrees. Homozygosity for eight previously unreported variants in transcribed sequences was detected by evaluating a library of 402,554 sequencing reads and was later confirmed by Sanger sequencing. Of these variants, six were determined to be polymorphisms in the Pakistani population, and one was in a noncoding gene that was subsequently excluded genetically from the DFNB79 linkage interval. The remaining variant was a nonsense mutation in a predicted gene, C9orf75, renamed TPRN. Evaluation of the other three DFNB79-linked families identified three additional frameshift mutations, for a total of four truncating alleles of this gene. Although TPRN is expressed in many tissues, immunolocalization of the protein product in the mouse cochlea shows prominent expression in the taper region of hair cell stereocilia. Consequently, we named the protein taperin.
Copyright 2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
_DFNB79-_Linked Pedigrees and Sequence Traces Showing Pathogenic Variants (A) Affected individual IV-4 from family PKDF741 was selected for NimbleGen capture and enrichment of genomic DNA from the DFNB79 locus and next-generation sequencing. Filled symbols in each pedigree represent affected individuals. A dot in the upper right corner of symbols for females (circles) and males (squares) indicates an individual whose DNA sequence of TPRN was determined via dideoxy-terminator chemistry. Bars below the DNA sequence indicate altered nucleotide(s). In family PKDF280, the sequence trace from an affected individual shows a homozygous 11 bp duplication. In an affected individual from family PKDF1129, dotted lines indicate the location of the 11 bp deletion. NCBI Reference Sequences (NM_001128228.1 for nucleotide change and NP_001121700 for amino acid change) were used to indicate positions of mutations in cDNA and protein, respectively. (B) The 11 bp deletion and 11 bp duplication found in affected members of families PKDF1129 and PKDF280, respectively, occur in sequence that is already duplicated in the wild-type.
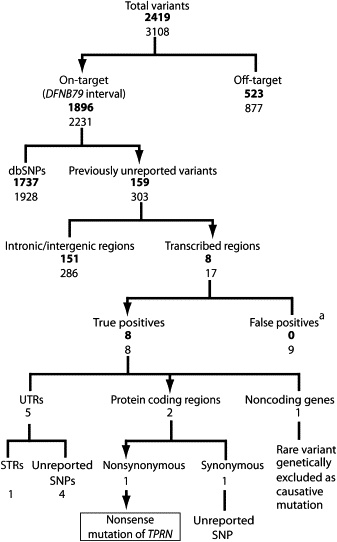
Figure 2
Summary of Variants Found by Next-Generation Sequencing Values in regular font are variants supported by at least three nonduplicate forward and reverse 454 reads or at least five unidirectional reads with a quality score greater then 20 (or 30 if the difference involved a pentamer or larger). Values in bold are restricted to those variants supported by at least 85% of the reads mapped at that location. Abbreviations are as follows: UTR, untranslated region; STR, short tandem repeat; SNP, single-nucleotide polymorphism. a False positives demonstrated by Sanger sequencing.
Figure 3
NimbleGen Probe Coverage and Sequencing Coverage across TPRN, GC Content and Locations of Four Pathogenic Variants Found in _DFNB79-_Linked Families Approximately 10 kb spanning TPRN in the 2.9 Mb DFNB79 interval is shown. Vertical lines correspond to GC content in a five-base window. The upper black bars depict probe coverage (tiled regions) of TPRN. Regions for which 454 sequence was obtained are shown in the lower set of bars. Note that in exon 1, a region of 878 bp that has 81% GC content was not sequenced even though it was tiled on the NimbleGen array. We determined the sequence of this GC-rich region from PCR products amplified from genomic DNA of family members of the four _DFNB79_-linked pedigrees by using dideoxy-terminator chemistry. Grey-colored thick bars, lines joining them, and the thin gray bar represent exons, introns, and the 3′ UTR, respectively, of TPRN. See Table S3 as well.
Figure 4
Conserved Synteny of a Part of Human Chromosome 9q with Mouse Chromosome 2 and Expression Profile of TPRN in Several Tissues (A) Part of human chromosome 9q, 17.71 Mb at the telomeric end, is syntenic to 14.62 Mb of mouse chromosome 2, which also includes 2.9 Mb of the DFNB79 linkage interval. Vertical lines (not drawn to scale) between the two chromosomal segments show the syntenic region, with five closely linked orthologous genes on each side of the DFNB79 gene. On the basis of conserved synteny and sequence comparison (see Figure 5), C430004E15Rik is the mouse ortholog of human TPRN. (B) Arrows indicate the position of the forward and reverse primer used to amplify the C430004E15Rik transcript from mouse multiple-tissue cDNA libraries. Expected cDNA and gDNA product sizes are 2.4 kb and 6.9 kb, respectively. Amplimers were sequence confirmed.
Figure 5
ClustalW Alignment of Predicted Amino Acid Sequences Encoded by Human TPRN and Its Mouse Ortholog C430004E15Rik Taperin (711 residues, Ensembl, ENSP00000387100) has 68% identity and 75% similarity with the orthologous mouse predicted protein of 749 amino acids deduced from genomic DNA and confirmed by sequencing cDNA. Residues located in the black-, gray-, and white-colored blocks are identical, similar and mismatched, respectively, and dots represent gaps. “M” on top of the black triangle is the translation start site predicted for human taperin (RefSeq, NP_001121700). Note that the additional N-terminal 61 residues are highly conserved in the mouse ortholog. Red triangles point to the first affected amino acid due to the one nonsense mutation and the three frameshift mutations segregating in the four _DFNB79_-linked pedigrees. The rectangle outlines the conservation of the human protein residues that were used as an immunogen for the commercially available antibody (Sigma-Aldrich) to taperin, which we used to immunostain mouse inner ear tissues. See also Figure S1 for a ClustalW alignment of taperin with multiple species.
Figure 6
Antibody Validation and Immunolocalization of Taperin in the Inner Ear Sensory Epithelia (A) COS7 cells transfected with an expression vector containing a cDNA construct for full-length mouse taperin epitope tagged with GFP. (B and C) Staining of GFP-taperin-transfected COS7 cells with taperin antibody. The antibody immunofluorescence signal (red) colocalizes with GFP-taperin. (D) COS7 cell transfected with full-length GFP-phostensin (green) and stained with taperin antibody (red). No colocalization signal is observed, revealing no crossreactivity of taperin antibody with phostensin. (E–H) Rhodamine-phalloidin (red) in the mouse organ of Corti and vestibular sensory epithelium highlights filamentous actin of stereocilia core, hair cell cuticular plates, as well as cytoskeleton of nonsensory cells. (E) An optical section of P7 mouse organ of Corti at the surface of the reticular lamina. Taperin (green) is localized at the base of each stereocilium of outer hair cells (OHCs) and inner hair cells (IHCs) at the top of the actin-rich cuticular plate. Inset shows a higher-resolution image of an OHC apical surface with taperin (green) localized near the base of each stereocilium from all three rows. Taperin immunoreactivity is also present in the supporting cells (SCs). The inner sulcus cells (ISCs) and other cell types outside of the sensory epithelium have weaker or no obvious taperin immunoreactivity. (F) Taperin (green) is present in the taper region at the base of each stereocilium (arrow) of vestibular hair cells (VHCs) in the sensory epithelium of mouse saccule. (G and H) Higher-resolution images of IHC stereocilia reveal distinct localization of taperin along the taper region near stereocilia bases at the apical surface of P7 (G) and P45 (H) hair cells (arrows). Scale bars in (C) and (D) represent 20 μm, and those in (E–H) represent 5 μm. See Figure S2 as well for a ClustalW alignment of human taperin and human phostensin.
Similar articles
- Mutations in TPRN cause a progressive form of autosomal-recessive nonsyndromic hearing loss.
Li Y, Pohl E, Boulouiz R, Schraders M, Nürnberg G, Charif M, Admiraal RJ, von Ameln S, Baessmann I, Kandil M, Veltman JA, Nürnberg P, Kubisch C, Barakat A, Kremer H, Wollnik B. Li Y, et al. Am J Hum Genet. 2010 Mar 12;86(3):479-84. doi: 10.1016/j.ajhg.2010.02.003. Epub 2010 Feb 18. Am J Hum Genet. 2010. PMID: 20170898 Free PMC article. - DFNB79: reincarnation of a nonsyndromic deafness locus on chromosome 9q34.3.
Khan SY, Riazuddin S, Shahzad M, Ahmed N, Zafar AU, Rehman AU, Morell RJ, Griffith AJ, Ahmed ZM, Riazuddin S, Friedman TB. Khan SY, et al. Eur J Hum Genet. 2010 Jan;18(1):125-9. doi: 10.1038/ejhg.2009.121. Eur J Hum Genet. 2010. PMID: 19603065 Free PMC article. - Progressive hearing loss and degeneration of hair cell stereocilia in taperin gene knockout mice.
Chen M, Wang Q, Zhu GH, Hu P, Zhou Y, Wang T, Lai RS, Xiao ZA, Xie DH. Chen M, et al. Biochem Biophys Res Commun. 2016 Oct 28;479(4):703-707. doi: 10.1016/j.bbrc.2016.09.148. Epub 2016 Sep 29. Biochem Biophys Res Commun. 2016. PMID: 27693694 - The c.42_52del11 mutation in TPRN and progressive hearing loss in a family from Pakistan.
Bashir R, Imtiaz A, Fatima A, Alam A, Naz S. Bashir R, et al. Biochem Genet. 2013 Jun;51(5-6):350-7. doi: 10.1007/s10528-013-9568-y. Epub 2013 Jan 23. Biochem Genet. 2013. PMID: 23340767 Free PMC article. - Mutations in TBC1D24, a gene associated with epilepsy, also cause nonsyndromic deafness DFNB86.
Rehman AU, Santos-Cortez RL, Morell RJ, Drummond MC, Ito T, Lee K, Khan AA, Basra MA, Wasif N, Ayub M, Ali RA, Raza SI; University of Washington Center for Mendelian Genomics; Nickerson DA, Shendure J, Bamshad M, Riazuddin S, Billington N, Khan SN, Friedman PL, Griffith AJ, Ahmad W, Riazuddin S, Leal SM, Friedman TB. Rehman AU, et al. Am J Hum Genet. 2014 Jan 2;94(1):144-52. doi: 10.1016/j.ajhg.2013.12.004. Am J Hum Genet. 2014. PMID: 24387994 Free PMC article.
Cited by
- Unveiling a novel GJB2 dominant K22T mutation in a Chinese family with hearing loss.
Ji H, Shu Y, Li H. Ji H, et al. Acta Biochim Biophys Sin (Shanghai). 2024 Jun 25;56(6):945-951. doi: 10.3724/abbs.2024064. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 38733163 Free PMC article. - Autosomal recessive non-syndromic hearing loss genes in Pakistan during the previous three decades.
Shadab M, Abbasi AA, Ejaz A, Ben-Mahmoud A, Gupta V, Kim HG, Vona B. Shadab M, et al. J Cell Mol Med. 2024 Apr;28(8):e18119. doi: 10.1111/jcmm.18119. J Cell Mol Med. 2024. PMID: 38534090 Free PMC article. Review. - Critical role of TPRN rings in the stereocilia for hearing.
Qi J, Tan F, Zhang L, Zhou Y, Zhang Z, Sun Q, Li N, Fang Y, Chen X, Wu Y, Zhong G, Chai R. Qi J, et al. Mol Ther. 2024 Jan 3;32(1):204-217. doi: 10.1016/j.ymthe.2023.11.011. Epub 2023 Nov 11. Mol Ther. 2024. PMID: 37952086 - Advanced Omics Techniques for Understanding Cochlear Genome, Epigenome, and Transcriptome in Health and Disease.
Tisi A, Palaniappan S, Maccarrone M. Tisi A, et al. Biomolecules. 2023 Oct 17;13(10):1534. doi: 10.3390/biom13101534. Biomolecules. 2023. PMID: 37892216 Free PMC article. Review. - The actin cytoskeleton in hair bundle development and hearing loss.
Park J, Bird JE. Park J, et al. Hear Res. 2023 Sep 1;436:108817. doi: 10.1016/j.heares.2023.108817. Epub 2023 May 26. Hear Res. 2023. PMID: 37300948 Free PMC article. Review.
References
- Scott H.S., Kudoh J., Wattenhofer M., Shibuya K., Berry A., Chrast R., Guipponi M., Wang J., Kawasaki K., Asakawa S. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat. Genet. 2001;27:59–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous