Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone - PubMed (original) (raw)
Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone
Mark B Meyer et al. J Steroid Biochem Mol Biol. 2010 Jul.
Abstract
The vitamin D receptor (VDR) mediates the actions of 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) in target cells and tissues by orchestrating the expression of gene networks responsible for vitamin D-induced phenotypes. The molecular mechanisms of these regulatory systems have been studied for decades under the principle that transcriptional regulation occurs near the transcriptional start site of the gene. However, this now appears to be an outdated view of transcriptional control. In this study, we examined the genome-wide chromatin immunoprecipitation on microarray (ChIP-chip) across pre-osteoblastic cells for VDR, retinoid X receptor (RXR), RNA polymerase II, and histone H4 acetylation (H4ac). We uncovered potential regulatory mechanisms for genes important to osteoblast biology as well as skeletal formation under the control of 1,25(OH)2D3. We found that VDR, along with RXR and H4ac, binds to distal regions 43% of the time; and within gene introns and exons 44%, leaving only 13% of activation at traditional promoter regions. Here, we briefly summarize our findings for all the VDR/RXR cis-acting transcriptional elements (VDR/RXR cistrome) in pre-osteoblastic cells, MC3T3-E1, provide a few examples of this dynamic control by VDR and 1,25(OH)2D3, and demonstrate that distal transcriptional control contributes to the majority of vitamin D3-mediated transcription.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures
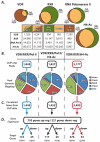
Figure 1
Summary of genome-wide ChIP-chip and gene expression studies reveal the VDR/RXR cistrome. A, Schematic venn diagram representations of the data displayed in the table below summarizing the number of peaks found with vehicle treatment (Veh/Input) and with 1,25(OH)2D3 treatment (1,25/Input) as well as Vehicle only, 1,25 only, and an overlap of regions from ChIP-chip data for VDR, RXR, RNA polymerase II (RNA Pol II) and histone H4 acetylation (H4ac). MC3T3-E1 cells treated for 3 hours prior to ChIP with vehicle or 100nM 1,25(OH)2D3. B, Overlapping peaks were tabulated for VDR/RXR/RNA Pol II (left column), VDR/RXR/RNA Pol II/H4ac (center column) and VDR/RXR/H4ac (right column). Peaks were mapped to their surrounding genes and categorized into intragenic (Intron or Exon), 5′near (within 5kb upstream of the 5′ end of gene), 3′near (within 5kb downstream of 3′ end of gene) or Distal (any region not within the gene or within 5kb of the gene at either end). C, the peaks were mapped to their closest surrounding gene and listed as correlated genes from ChIP-chip data. D, gene expression analysis was performed on MC3T3-E1 cells for 24 hours with either vehicle or 100nM 1,25(OH)2D3. These genes were then cross referenced with the genes that were associated with the ChIP-chip peaks and displayed as either up- or down-regulated genes.
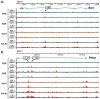
Figure 2
ChIP-chip data for genes important for skeletal biology reveal novel enhancers at the promoter as well as distal to the TSS. A, the genomic location for the Spp1 (Opn) gene are shown for chromosome 5 with genomic base pairs given in kilobases (k). ChIP-chip data are listed for each antibody in the basal (Veh/Input) or activated (1,25/Input) state and are displayed as log 2 ratios (log2R). Antibodies used are VDR (blue), RXR (green), RNA Pol II (orange), H4ac (brown). Statistically significant peaks were called and are highlighted in red as described in Materials and Methods. B, the same analysis is shown for the Cyp24a1 gene. Putative regulatory regions discovered are highlighted with their position relative to the transcriptional start site of the gene.
Figure 3
Transcriptional control can occur within intragenic regions at introns and exons. A, the genomic location for the Rarβ gene are shown for chromosome 5 with genomic base pairs given in kilobases (k). ChIP-chip data are listed for each antibody in the basal (Veh/Input) or activated (1,25/Input) state and are displayed as log 2 ratios (log2R). Antibodies used are VDR (blue), RXR (green), RNA Pol II (orange), H4ac (brown). Statistically significant peaks were called and are highlighted in red as described in Materials and Methods. B, the same analysis is shown for the Prcka (Protein Kinase Cα) gene. Putative regulatory regions discovered are highlighted with their position relative to the transcriptional start site of the gene.
Similar articles
- Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3.
Zella LA, Kim S, Shevde NK, Pike JW. Zella LA, et al. Mol Endocrinol. 2006 Jun;20(6):1231-47. doi: 10.1210/me.2006-0015. Epub 2006 Feb 23. Mol Endocrinol. 2006. PMID: 16497728 - The vitamin D hormone and its nuclear receptor: molecular actions and disease states.
Haussler MR, Haussler CA, Jurutka PW, Thompson PD, Hsieh JC, Remus LS, Selznick SH, Whitfield GK. Haussler MR, et al. J Endocrinol. 1997 Sep;154 Suppl:S57-73. J Endocrinol. 1997. PMID: 9379138 Review. - New understanding of the molecular mechanism of receptor-mediated genomic actions of the vitamin D hormone.
Haussler MR, Jurutka PW, Hsieh JC, Thompson PD, Selznick SH, Haussler CA, Whitfield GK. Haussler MR, et al. Bone. 1995 Aug;17(2 Suppl):33S-38S. doi: 10.1016/8756-3282(95)00205-r. Bone. 1995. PMID: 8579895 Review.
Cited by
- In vivo contribution of Cyp24a1 promoter vitamin D response elements.
Meyer MB, Lee SM, Towne JM, Cichanski SR, Kaufmann M, Jones G, Pike JW. Meyer MB, et al. bioRxiv [Preprint]. 2024 Aug 24:2024.08.23.609393. doi: 10.1101/2024.08.23.609393. bioRxiv. 2024. PMID: 39229197 Free PMC article. Updated. Preprint. - FGF23 gene regulation by 1,25-dihydroxyvitamin D: opposing effects in adipocytes and osteocytes.
Kaneko I, Saini RK, Griffin KP, Whitfield GK, Haussler MR, Jurutka PW. Kaneko I, et al. J Endocrinol. 2015 Sep;226(3):155-66. doi: 10.1530/JOE-15-0225. Epub 2015 Jul 6. J Endocrinol. 2015. PMID: 26148725 Free PMC article. - The osteoblast to osteocyte transition: epigenetic changes and response to the vitamin D3 hormone.
St John HC, Bishop KA, Meyer MB, Benkusky NA, Leng N, Kendziorski C, Bonewald LF, Pike JW. St John HC, et al. Mol Endocrinol. 2014 Jul;28(7):1150-65. doi: 10.1210/me.2014-1091. Epub 2014 May 30. Mol Endocrinol. 2014. PMID: 24877565 Free PMC article. - 1,25-dihydroxyvitamin D(3) regulation of fibroblast growth factor-23 expression in bone cells: evidence for primary and secondary mechanisms modulated by leptin and interleukin-6.
Saini RK, Kaneko I, Jurutka PW, Forster R, Hsieh A, Hsieh JC, Haussler MR, Whitfield GK. Saini RK, et al. Calcif Tissue Int. 2013 Apr;92(4):339-53. doi: 10.1007/s00223-012-9683-5. Epub 2012 Dec 22. Calcif Tissue Int. 2013. PMID: 23263654 Free PMC article. - Vitamin D in follicular development and oocyte maturation.
Xu F, Wolf S, Green O, Xu J. Xu F, et al. Reproduction. 2021 May 5;161(6):R129-R137. doi: 10.1530/REP-20-0608. Reproduction. 2021. PMID: 33835047 Free PMC article. Review.
References
- Jones G, Strugnell SA, DeLuca HF. Current understanding of the molecular actions of vitamin D. Physiol Rev. 1998;78(4):1193–1231. - PubMed
- Jurutka PW, Whitfield GK, Hsieh JC, Thompson PD, Haussler CA, Haussler MR. Molecular nature of the vitamin D receptor and its role in regulation of gene expression. Rev Endocr Metab Disord. 2001;2(2):203–216. - PubMed
- Malloy P, Pike J, Feldman D. The vitamin D receptor and the syndrome of hereditary 1,25-dihydroxyvitamin D-resistant rickets. Endocr Rev. 1999;20(2):156–188. - PubMed
- Mangelsdorf D, Evans R. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–850. - PubMed
- Malloy PJ, Pike JW, Feldman D. The vitamin D receptor and the syndrome of hereditary 1,25-dihydroxyvitamin D-resistant rickets. Endocr Rev. 1999;20(2):156–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK072281/DK/NIDDK NIH HHS/United States
- R01 DK073995/DK/NIDDK NIH HHS/United States
- R01 DK072281-05/DK/NIDDK NIH HHS/United States
- R01 DK074993/DK/NIDDK NIH HHS/United States
- R01 AR045173-05/AR/NIAMS NIH HHS/United States
- R01 AR045173/AR/NIAMS NIH HHS/United States
- DK03228/DK/NIDDK NIH HHS/United States
- R01 DK073995-03/DK/NIDDK NIH HHS/United States
- R01 DK074993-04/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases