NFATc4 is negatively regulated in miR-133a-mediated cardiomyocyte hypertrophic repression - PubMed (original) (raw)
NFATc4 is negatively regulated in miR-133a-mediated cardiomyocyte hypertrophic repression
Qi Li et al. Am J Physiol Heart Circ Physiol. 2010 May.
Abstract
Activation of NFAT (nuclear factor of activated T cells)-mediated hypertrophic signaling is a major regulatory response to hypertrophic stimuli. A recent study unveiled potential regulatory roles for microRNA-133a (miR-133a) in cardiac hypertrophy. To date, however, no connection has been made between miR-133a and NFAT signaling. In this study, we determined that NFATc4, a hypertrophy-associated mediator, is negatively regulated by miR-133a. Two conserved base-pairing sites between the NFATc4 3'-untranslated region (UTR) and miR-133a were verified. Mutation of these sites in the NFATc4 3'-UTR completely blocked the negative effect of miR-133a on NFATc4, suggesting that NFATc4 is a direct target for miR-133a regulation. Using a gain-of-function approach, we demonstrate that miR-133 significantly reduces the endogenous level of, as well as the hypertrophic stimulus-mediated increase in, NFATc4 gene expression. This latter effect of miR-133a on NFATc4 gene expression was coincided with an attenuated cardiomyocyte hypertrophy induced by an alpha-adrenergic receptor agonist. Conversely, cells treated with miR-133a inhibitor resulted in an increase in NFATc4 expression level. Application of miR-133a had no apparent effect on NFATc4 nuclear localization. We conclude that the negative regulation of NFATc4 expression contributes to miR-133a-mediated hypertrophic repression.
Figures
Fig. 1.
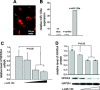
Prediction of two putative targeting sites of micro-RNA (miR)-133a in the 3′-untranslated region (UTR) of nuclear factor of activated T cell (NFAT) c4. A: schematic of exon 10 of NFATc4, including 3′-UTR. B: alignment of mouse miR-133a sequence with the 3′-UTRs of NFATc4 among different species. Matched nucleotides were highlighted in bold. Two highly conserved 3′-UTR sites were recognized.
Fig. 2.
Analysis of the NFATc4–3′-UTR by luciferase activity assay. A: schematic presentation of the luciferase assay. The NFATc4–3′-UTR containing two miR-133a targeting sites was fused with the reporter gene luciferase. B: addition of the miR-133a mimic attenuated NFATc4–3′-UTR reporter gene activity. Mutation of miR-133a putative target sites blocked the repressive effect of miR-133a on the target, suggesting NFATc4 as a miR-133a target gene. The data in each group represent the average of nine measurements.
Fig. 3.
Effect of miR-133a mimic and inhibitor on the expression of NFATc4. Treatment of C2C12 cells with chemically modified miR-133a mimic reduced the levels of NFATc4 mRNA and protein. The converse results were observed when cells were treated with miR-133a inhibitor. A: quantitative polymerase chain reaction (qPCR) analysis. Each group contained six measurements. B: the expression level of NFATc4 protein was assessed by Western blot and semiquantified by densitometric analysis with six measurements in each group. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Fig. 4.
Generation of the adenovirus expressing miR-133a (v-miR-133a) and assessment of miR-133a-mediated NFATc4 silencing. A: red fluorescent protein was coexpressed as a marker along with the miR expression. B: qPCR analysis revealed a high level of miR-133a expressed by v-miR-133a. C and D: dose-dependent silencing of NFATc4 by v-miR-133a was assessed in C2C12 cells by qPCR and Western blot analysis, respectively. v-control, Control adenovirus expressing RFP. The data in each group represent the average of nine measurements, except for B and D, which contains four measurements.
Fig. 5.
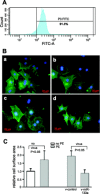
miR-133a treatment attenuates phenylephrine (PE)-induced cardiomyocyte hypertrophy. A: flow cytometry demonstrated 91.3% purity in neonatal rat cardiomyocytes. B: representative immunostaining of cardiomyocytes: control cardiomyocytes (a); PE-treated cardiomyocytes (b); PE-treated cardiomyocytes with control virus (c); PE-treated cardiomyocytes with v-miR-133a (d). C: morphometric analysis of cardiomyocytes. Size of cardiomyocytes, as evaluated by surface area, was significantly increased by PE treatment. This size increase was blocked by v-miR-133a. Cardiomyocytes were detected by cardiac-specific troponin I antibodies (green), followed by diamidino-2-phenylindole staining for nuclei (blue). At least 50 cardiomyocytes were randomly chosen for cell surface area measurement.
Fig. 6.
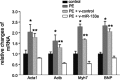
PE-mediated hypertrophic responses in cardiomyocytes were eliminated by miR-133a. The hypertrophic response was demonstrated in robust increases in various stress markers. The changes of the stress marker mRNAs were quantified by qPCR analysis. P < 0.05 compared with *controls or **PE + v-miR-133a groups. The data in each group represent the average of nine measurements. BNP, brain natriuretic peptide.
Fig. 7.
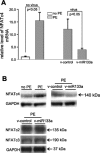
Silencing of NFATc4 gene expression by miR-133a. In response to PE and miR-133a treatments, expression of NFATc4 was assessed by qPCR (A) and Western blot analysis (B), respectively. Robust increases in NFATc4 mRNA and protein were observed after the PE treatment, whereas the increases were attenuated by infection of the v-miR-133a. Expression levels of NFATc2 and NFATc3 did not significantly change on v-miR-133a treatment. The data in each group in A represent the average of nine measurements.
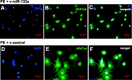
Fig. 8.
Application of miR-133a had no effect on NFATc4 nuclear localization. Immunostaining with antibody specific for NFATc4 was performed in neonatal rat ventricular myocytes. Expression of NFATc4 (green) was observed in both nucleus and cytoplasm. No significant changes of NFATc4 cellular distribution were found between the v-miR-133a (A–C) and v-control virus (D–F) treatment groups.
Similar articles
- MicroRNA-29a-3p attenuates ET-1-induced hypertrophic responses in H9c2 cardiomyocytes.
Li M, Wang N, Zhang J, He HP, Gong HQ, Zhang R, Song TF, Zhang LN, Guo ZX, Cao DS, Zhang TC. Li M, et al. Gene. 2016 Jul 1;585(1):44-50. doi: 10.1016/j.gene.2016.03.015. Epub 2016 Mar 15. Gene. 2016. PMID: 26992639 - [Effects of hydrogen sulfide (H2S) on cardiac hypertrophy and miRNA-133a-mediated Ca2+/calcineurin/NFATc4 signal pathway in rats].
Wu Y, Guo YY, Zhang YY, Zhang Y. Wu Y, et al. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2018 Jan 8;34(1):29-34. doi: 10.12047/j.cjap.5492.2018.009. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2018. PMID: 29926655 Chinese. - Mitochondrial Disruption Is Involved in the Effect of Nuclear Factor of Activated T cells, Cytoplasmic 4 on Aggravating Cardiomyocyte Hypertrophy.
Liu X, Gao S, Gao H, Jiang X, Wei Q. Liu X, et al. J Cardiovasc Pharmacol. 2021 May 1;77(5):557-569. doi: 10.1097/FJC.0000000000000986. J Cardiovasc Pharmacol. 2021. PMID: 33951694 - MicroRNAs in cardiomyocyte development.
Malizia AP, Wang DZ. Malizia AP, et al. Wiley Interdiscip Rev Syst Biol Med. 2011 Mar-Apr;3(2):183-90. doi: 10.1002/wsbm.111. Wiley Interdiscip Rev Syst Biol Med. 2011. PMID: 21305703 Free PMC article. Review. - Recent knowledge of NFATc4 in oncogenesis and cancer prognosis.
Zhong QH, Zha SW, Lau ATY, Xu YM. Zhong QH, et al. Cancer Cell Int. 2022 Jun 13;22(1):212. doi: 10.1186/s12935-022-02619-6. Cancer Cell Int. 2022. PMID: 35698138 Free PMC article. Review.
Cited by
- Mutual antagonism between IP(3)RII and miRNA-133a regulates calcium signals and cardiac hypertrophy.
Drawnel FM, Wachten D, Molkentin JD, Maillet M, Aronsen JM, Swift F, Sjaastad I, Liu N, Catalucci D, Mikoshiba K, Hisatsune C, Okkenhaug H, Andrews SR, Bootman MD, Roderick HL. Drawnel FM, et al. J Cell Biol. 2012 Nov 26;199(5):783-98. doi: 10.1083/jcb.201111095. Epub 2012 Nov 19. J Cell Biol. 2012. PMID: 23166348 Free PMC article. - Comparative Analyses of MicroRNA Microarrays during Cardiogenesis: Functional Perspectives.
Bonet F, Hernandez-Torres F, Esteban FJ, Aranega A, Franco D. Bonet F, et al. Microarrays (Basel). 2013 Apr 3;2(2):81-96. doi: 10.3390/microarrays2020081. Microarrays (Basel). 2013. PMID: 27605182 Free PMC article. Review. - MicroRNAs in Hypertrophic, Arrhythmogenic and Dilated Cardiomyopathy.
Chiti E, Paolo MD, Turillazzi E, Rocchi A. Chiti E, et al. Diagnostics (Basel). 2021 Sep 19;11(9):1720. doi: 10.3390/diagnostics11091720. Diagnostics (Basel). 2021. PMID: 34574061 Free PMC article. Review. - Regulation of nuclear factor of activated T cells (NFAT) and downstream myogenic proteins during dehydration in the African clawed frog.
Zhang Y, English SG, Storey KB. Zhang Y, et al. Mol Biol Rep. 2018 Oct;45(5):751-761. doi: 10.1007/s11033-018-4214-8. Epub 2018 Jun 19. Mol Biol Rep. 2018. PMID: 29923155 - MicroRNAs in heart development.
Espinoza-Lewis RA, Wang DZ. Espinoza-Lewis RA, et al. Curr Top Dev Biol. 2012;100:279-317. doi: 10.1016/B978-0-12-387786-4.00009-9. Curr Top Dev Biol. 2012. PMID: 22449848 Free PMC article. Review.
References
- Ambros V. The functions of animal microRNAs. Nature 431: 350–355, 2004 - PubMed
- Bao Y, Li R, Jiang J, Cai B, Gao J, Le K, Zhang F, Chen S, Liu P. Activation of peroxisome proliferator-activated receptor gamma inhibits endothelin-1-induced cardiac hypertrophy via the calcineurin/NFAT signaling pathway. Mol Cell Biochem 317: 189–196, 2008 - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, function. Cell 116: 281–297, 2004 - PubMed
- Bushdid PB, Osinska H, Waclaw RR, Molkentin JD, Yutzey KE. NFATc3 and NFATc4 are required for cardiac development and mitochondrial function. Circ Res 92: 1305–1313, 2003 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 HL094844-01A2/HL/NHLBI NIH HHS/United States
- R21 HL094844/HL/NHLBI NIH HHS/United States
- K02 HL098956/HL/NHLBI NIH HHS/United States
- R01 HL102314-01/HL/NHLBI NIH HHS/United States
- R01 HL102314/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous