Inflammatory stimuli regulate caspase substrate profiles - PubMed (original) (raw)
Inflammatory stimuli regulate caspase substrate profiles
Nicholas J Agard et al. Mol Cell Proteomics. 2010 May.
Abstract
The inflammatory caspases, human caspases-1, -4, and -5, proteolytically modulate diverse physiological outcomes in response to proinflammatory signals. Surprisingly, only a few substrates are known for these enzymes, including other caspases and the interleukin-1 family of cytokines. To more comprehensively characterize inflammatory caspase substrates, we combined an enzymatic N-terminal enrichment method with mass spectrometry-based proteomics to identify newly cleaved proteins. Analysis of THP-1 monocytic cell lysates treated with recombinant purified caspases identified 82 putative caspase-1 substrates, three putative caspase-4 substrates, and no substrates for caspase-5. By contrast, inflammatory caspases activated in THP-1 cells by mimics of gout (monosodium urate), bacterial infection (lipopolysaccharide and ATP), or viral infection (poly(dA.dT)) were found to cleave only 27, 16, and 22 substrates, respectively. Quantitative stable isotope labeling with amino acids in cell culture (SILAC) comparison of these three inflammatory stimuli showed that they induced largely overlapping substrate profiles but different extents of proteolysis. Interestingly, only half of the cleavages found in response to proinflammatory stimuli were contained within our set of 82 in vitro cleavage sites. These data provide the most comprehensive set of caspase-1-cleaved products reported to date and indicate that caspases-4 and -5 have far fewer substrates. Comparisons between the in vitro and in vivo data highlight the importance of localization in regulating inflammatory caspase activity. Finally, our data suggest that inducers of inflammation may subtly alter caspase-1 substrate profiles.
Figures
Fig. 1.
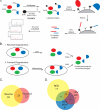
Degradomics approach for identifying caspase-cleaved peptides. A, to identify cleavage sites, lysates are N-terminally biotinylated with subtiligase and TEVest2. The labeled proteins are captured on streptavidin beads, trimmed to a single peptide with trypsin, and released from the beads with TEV protease. The resulting labeled peptides are identified by LC/MS/MS. B, two modes are used for identifying caspase-cleaved peptides. i, in reverse degradomics, purified caspase is added exogenously to cellular lysates, generating caspase cleavage events. ii, in forward degradomics, inflammatory (or apoptotic) stimuli induce caspase activation in the cell or conditioned media. These stimulated cells are then lysed and analyzed. C, the overlap of forward and reverse degradomics data sets. D, the overlap between forward degradomics data sets for three different inflammatory stimuli.
Fig. 2.
Caspase-1-cleaved peptides show propensity for hydrophobic residues at P4. A, sequences surrounding the cleavage site of aspartyl-cleaved protein (P1 = Asp) were aligned and analyzed in a sequence logo. Amino acid letter sizes correspond to the frequency; colors correspond to the side chain functionality as follows: acidic (Asp and Glu), red; hydrophobic (Phe, His, Trp, and Tyr), light blue; aliphatic (Leu, Ile, Met, Val, and Pro), dark blue; small (Ala, Gly, Ser, and Thr), green; and other (Cys, Asn, Lys, Gln, and Arg), black. B, SILAC comparisons between caspase-1-treated and untreated lysates were graphed on a double log plot. Aspartyl-cleaved peptides are color-coded according to their P4 residue: putative apoptotic substrates (Asp and Glu), putative inflammatory substrates (Leu, Ile, Met, Val, Pro, Phe, His, Tyr, and Trp), and unknown (Ala, Gly, Ser, and Thr). Note that the putative inflammatory substrates have a high light/heavy ratio, suggesting that they are produced by the addition of caspase-1. Substrates that were selected for further biochemical examination are circled. C, bar graph representation of the caspase-cleaved peptides in B grouped by SILAC ratio. Note that the caspase-derived peptides with the highest light/heavy ratios contained large hydrophobic residues in P4, consistent with caspase-1 specificity.
Fig. 3.
In vitro analyses of caspase-1-cleaved substrates show wide variation in rate and selectivity. A, top, individual substrates were expressed by IVT and treated with 50 n
m
caspase-1 (GSDMD) or 200 n
m
caspase-1 (all others) (representative data is shown). At the indicated time points, aliquots were diluted with SDS loading buffer and stored flash frozen prior to analysis by SDS-PAGE. Arrows indicate the masses of cleaved products. Bottom, substrates were treated with 50 n
m
(GSDMD) or 200 n
m
(all others) caspases-1–9 and incubated for 5 min (GSDMD) or 90 min (all others) prior to analysis by SDS-PAGE. B, fluorescence signals from A were integrated and plotted. Lines were fit to standard first order decay, f = _f_0 + A_0_e_−_kt where f is fluorescence intensity, _f_0 is background fluorescence, _A_0 is the fluorescence intensity of the intact protein, k is the rate of the reaction, and t is time. C, apparent _k_cat/Km was calculated for each substrate by assuming [substrate] ≪ Km and applying k = (_k_cat/Km)app × [caspase-1]. D, the caspase screen described in A was applied to SYAP1 D281A. E, caspase specificity was analyzed by integrating the fluorescence intensity of the full-length protein. +++, >60% reduction in intensity compared with no caspase control; ++, >40% reduction; +, >20% reduction or visual evidence of a cleaved product; −, >80% reduction of caspase intensity and no visible products. AU, arbitrary units; Casp, caspase; ZYX, zyxin.
Fig. 4.
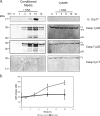
DNA transfection mediates caspase activation and substrate release. A, THP-1 cells were transfected with or without DNA for the indicated times. Cell extracts and conditioned media were prepared and probed by immunoblot for the presence of caspases-1, -3, and -7 and IL-1β. B, release of lactate dehydrogenase (LDH) was assessed for THP-1 cells under the same conditions. Casp, caspase; AU, arbitrary units.
Similar articles
- Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes.
Lamkanfi M, Kanneganti TD, Van Damme P, Vanden Berghe T, Vanoverberghe I, Vandekerckhove J, Vandenabeele P, Gevaert K, Núñez G. Lamkanfi M, et al. Mol Cell Proteomics. 2008 Dec;7(12):2350-63. doi: 10.1074/mcp.M800132-MCP200. Epub 2008 Jul 30. Mol Cell Proteomics. 2008. PMID: 18667412 Free PMC article. - Quantitative MS-based enzymology of caspases reveals distinct protein substrate specificities, hierarchies, and cellular roles.
Julien O, Zhuang M, Wiita AP, O'Donoghue AJ, Knudsen GM, Craik CS, Wells JA. Julien O, et al. Proc Natl Acad Sci U S A. 2016 Apr 5;113(14):E2001-10. doi: 10.1073/pnas.1524900113. Epub 2016 Mar 22. Proc Natl Acad Sci U S A. 2016. PMID: 27006500 Free PMC article. - Transcription factor AP-2alpha is preferentially cleaved by caspase 6 and degraded by proteasome during tumor necrosis factor alpha-induced apoptosis in breast cancer cells.
Nyormoi O, Wang Z, Doan D, Ruiz M, McConkey D, Bar-Eli M. Nyormoi O, et al. Mol Cell Biol. 2001 Aug;21(15):4856-67. doi: 10.1128/MCB.21.15.4856-4867.2001. Mol Cell Biol. 2001. PMID: 11438643 Free PMC article. - Inflammatory caspase substrate specificities.
Exconde PM, Bourne CM, Kulkarni M, Discher BM, Taabazuing CY. Exconde PM, et al. mBio. 2024 Jul 17;15(7):e0297523. doi: 10.1128/mbio.02975-23. Epub 2024 Jun 5. mBio. 2024. PMID: 38837391 Free PMC article. Review. - Caspases and their substrates.
Julien O, Wells JA. Julien O, et al. Cell Death Differ. 2017 Aug;24(8):1380-1389. doi: 10.1038/cdd.2017.44. Epub 2017 May 12. Cell Death Differ. 2017. PMID: 28498362 Free PMC article. Review.
Cited by
- ASC filament formation serves as a signal amplification mechanism for inflammasomes.
Dick MS, Sborgi L, Rühl S, Hiller S, Broz P. Dick MS, et al. Nat Commun. 2016 Jun 22;7:11929. doi: 10.1038/ncomms11929. Nat Commun. 2016. PMID: 27329339 Free PMC article. - Interactome-wide analysis identifies end-binding protein 1 as a crucial component for the speck-like particle formation of activated absence in melanoma 2 (AIM2) inflammasomes.
Wang LJ, Hsu CW, Chen CC, Liang Y, Chen LC, Ojcius DM, Tsang NM, Hsueh C, Wu CC, Chang YS. Wang LJ, et al. Mol Cell Proteomics. 2012 Nov;11(11):1230-44. doi: 10.1074/mcp.M112.020594. Epub 2012 Aug 6. Mol Cell Proteomics. 2012. PMID: 22869553 Free PMC article. - Early Exercise Protects against Cerebral Ischemic Injury through Inhibiting Neuron Apoptosis in Cortex in Rats.
Zhang P, Zhang Y, Zhang J, Wu Y, Jia J, Wu J, Hu Y. Zhang P, et al. Int J Mol Sci. 2013 Mar 15;14(3):6074-89. doi: 10.3390/ijms14036074. Int J Mol Sci. 2013. PMID: 23502470 Free PMC article. - Yersinia Type III-Secreted Effectors Evade the Caspase-4 Inflammasome in Human Cells.
Zhang J, Brodsky IE, Shin S. Zhang J, et al. bioRxiv [Preprint]. 2023 Jun 16:2023.01.24.525473. doi: 10.1101/2023.01.24.525473. bioRxiv. 2023. PMID: 36747770 Free PMC article. Updated. Preprint.
References
- Brasier A. R. (2006) The NF-kappaB regulatory network. Cardiovasc. Toxicol 6, 111–130 - PubMed
- Müntz K. (2007) Protein dynamics and proteolysis in plant vacuoles. J. Exp. Bot 58, 2391–2407 - PubMed
- Backes B. J., Harris J. L., Leonetti F., Craik C. S., Ellman J. A. (2000) Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat. Biotechnol 18, 187–193 - PubMed
- Baggio R., Burgstaller P., Hale S. P., Putney A. R., Lane M., Lipovsek D., Wright M. C., Roberts R. W., Liu R., Szostak J. W., Wagner R. W. (2002) Identification of epitope-like consensus motifs using mRNA display. J. Mol. Recognit 15, 126–134 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- S10 RR015804/RR/NCRR NIH HHS/United States
- R01 GM081051/GM/NIGMS NIH HHS/United States
- P41RR001614/RR/NCRR NIH HHS/United States
- F32 AI077177-03/AI/NIAID NIH HHS/United States
- F32 AI077177/AI/NIAID NIH HHS/United States
- R01 AI070292/AI/NIAID NIH HHS/United States
- P41 RR001614/RR/NCRR NIH HHS/United States
- F32AI077177/AI/NIAID NIH HHS/United States
- RR015804/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources