Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome - PubMed (original) (raw)
Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome
Juan M Vaquerizas et al. PLoS Genet. 2010.
Abstract
Transcriptional regulation is one of the most important processes for modulating gene expression. Though much of this control is attributed to transcription factors, histones, and associated enzymes, it is increasingly apparent that the spatial organization of chromosomes within the nucleus has a profound effect on transcriptional activity. Studies in yeast indicate that the nuclear pore complex might promote transcription by recruiting chromatin to the nuclear periphery. In higher eukaryotes, however, it is not known whether such regulation has global significance. Here we establish nucleoporins as a major class of global regulators for gene expression in Drosophila melanogaster. Using chromatin-immunoprecipitation combined with microarray hybridisation, we show that Nup153 and Megator (Mtor) bind to 25% of the genome in continuous domains extending 10 kb to 500 kb. These Nucleoporin-Associated Regions (NARs) are dominated by markers for active transcription, including high RNA polymerase II occupancy and histone H4K16 acetylation. RNAi-mediated knock-down of Nup153 alters the expression of approximately 5,700 genes, with a pronounced down-regulatory effect within NARs. We find that nucleoporins play a central role in coordinating dosage compensation-an organism-wide process involving the doubling of expression of the male X chromosome. NARs are enriched on the male X chromosome and occupy 75% of this chromosome. Furthermore, Nup153-depletion abolishes the normal function of the male-specific dosage compensation complex. Finally, by extensive 3D imaging, we demonstrate that NARs contribute to gene expression control irrespective of their sub-nuclear localization. Therefore, we suggest that NAR-binding is used for chromosomal organization that enables gene expression control.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
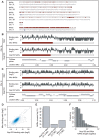
Figure 1. Nup153 and Megator bind the Drosophila genome on a large scale.
(A) Karyotype representation of the Drosophila genome; the upper track depicts the occurrence of high-density nucleoporin-binding in SL-2 cells and the lower track shows the location of annotated genes. Termed Nucleoporin Associated Regions (NARs), high-density binding occurs across 25% of the genome and there is particularly high occupancy on the male X chromosome. (B) Magnified view of Nup153 and Mtor-binding on chromosome 3L. For each nucleoporin, the upper track displays the processed ChIP/input profile and the lower track colours the sections identified as NARs. Note that Nup153 and Mtor show very similar patterns of binding. (C) Magnified view of nucleoporin-binding and NAR occurrence on chromosome X. There is much denser binding on this chromosome compared with autosomes. (D) Smoothed scatter plot displaying the ChIP/input binding ratios for Nup153 and Mtor (r = 0.77). (E) Barplot representing the overlap in NARs defined by Nup153 and Mtor binding profiles. (F) Histogram of Nup153 and Mtor NAR length distributions.
Figure 2. NARs define transcriptionally active regions of the genome.
Genome-track view of 1Mb section on chromosome 2L. NARs are enriched for transcribed genes compared with non-NARs (gene expression track; green shading), and a large proportion of genes are down-regulated upon Nup153-depletion (Nup153 RNAi track; red shading). NARs also align with markers of a transcriptionally active chromatin structure (H4K14Ac, MOF and PolII tracks; grey shading), but exclude markers for inactive chromatin (lamin, H3K27me3; grey shading).
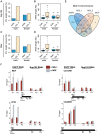
Figure 3. Male X chromosome is especially enriched for NARs.
Percentages of NAR occupancy on male and female autosomes and X chromosome for (A) Nup153 and (C) Mtor. In males, NARs are particularly enriched on the X chromosome compared with autosomes, whereas NARs occur evenly throughout in females. NAR length distributions for (B) Nup153 and (D) Mtor. NARs are much longer on the male X chromosome. (E) Overlap between NARs and MSL1-, MSL3- and MOF-binding; numbers represent gene counts. (F) Effect of Nup153-depletion on MSL1- (red shading) and MOF-binding (grey shading) to four X-chromosomal target loci. DNA prepared from cells treated with EGFP (control) or Nup153 dsRNA was immunoprecipitated and analysed by qPCR using primers for the beginning (P1), middle (P2) and end (P3) of genes. Error bars represent the standard deviation in measurements from three replicate experiments. Recovered DNA is shown as a percentage of input DNA.
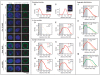
Figure 4. Nup153 and Mtor define NARs both at the periphery and the interior of the nucleus.
(A) Representative images of single confocal sections of nuclei containing the FISH signal (red) over DAPI (blue) and immunostained lamin (green). Target genomic regions include a lamin-bound gene (L105), NAR (T4, T15, T7, T11, T9, T13) and non-NAR loci (N1, N2). Probability density plots show the distribution of distance measurements between the FISH signal and the closest point on the nuclear boundary. Simulated nuclei show the ideal distributions for FISH targets located at the (B) periphery and (C) interior. Distances range from 0 at the boundary and 1.0 at the centroid of the nucleus. The grey background represents the theoretical 30% limit for a peripherally localised FISH signal. Observed distributions of in vivo controls for (D) peripherally localised L105 and (E) non-peripherally localised N2; the broad spread compared with simulations indicate that the loci display dynamic behaviour in their positioning within the nucleus. (F, G) Predominantly peripheral loci (T4, T15) have distributions that are similar to L105 (shown in yellow), whereas (H, I) predominantly non-peripheral loci (N1, T11) have very different distributions. Aggregate distributions for all NAR targets on (J) the male X chromosome and (L) autosomes, and all non-NAR targets on (K) the male X and (M) autosomes. Targets on the X chromosome are peripherally localised compared with autosomal ones.
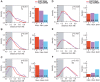
Figure 5. Peripheral localisation is dependent on Nup153.
Probability density distributions of distance measurements for mock treated (red) and Nup153-depleted cells (purple). Histograms depict the proportion of nuclei for which the FISH signal is located within the 30% distance threshold (DAPI in blue). (A-C) NAR targets on the male X chromosome (T4, T7, T5) relocalise to the interior upon treatment, indicating that peripheral localisation is dependent on Nup153. (D-F) NAR and non-NAR targets at the interior remain unaffected upon Nup153-depletion.
Similar articles
- Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex.
Kalverda B, Fornerod M. Kalverda B, et al. Cell Cycle. 2010 Dec 15;9(24):4812-7. doi: 10.4161/cc.9.24.14328. Epub 2010 Dec 15. Cell Cycle. 2010. PMID: 21150273 - Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila.
Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A. Mendjan S, et al. Mol Cell. 2006 Mar 17;21(6):811-23. doi: 10.1016/j.molcel.2006.02.007. Mol Cell. 2006. PMID: 16543150 - Correct dosage of X chromosome transcription is controlled by a nuclear pore component.
Aleman JR, Kuhn TM, Pascual-Garcia P, Gospocic J, Lan Y, Bonasio R, Little SC, Capelson M. Aleman JR, et al. Cell Rep. 2021 Jun 15;35(11):109236. doi: 10.1016/j.celrep.2021.109236. Cell Rep. 2021. PMID: 34133927 Free PMC article. - Dosage Compensation of the X Chromosome: A Complex Epigenetic Assignment Involving Chromatin Regulators and Long Noncoding RNAs.
Samata M, Akhtar A. Samata M, et al. Annu Rev Biochem. 2018 Jun 20;87:323-350. doi: 10.1146/annurev-biochem-062917-011816. Epub 2018 Apr 18. Annu Rev Biochem. 2018. PMID: 29668306 Review. - The right dose for every sex.
Mendjan S, Akhtar A. Mendjan S, et al. Chromosoma. 2007 Apr;116(2):95-106. doi: 10.1007/s00412-006-0089-x. Epub 2006 Nov 24. Chromosoma. 2007. PMID: 17124606 Free PMC article. Review.
Cited by
- Multiscale dynamics in nucleocytoplasmic transport.
Grünwald D, Singer RH. Grünwald D, et al. Curr Opin Cell Biol. 2012 Feb;24(1):100-6. doi: 10.1016/j.ceb.2011.11.011. Epub 2011 Dec 22. Curr Opin Cell Biol. 2012. PMID: 22196930 Free PMC article. Review. - Core Components of the Nuclear Pore Bind Distinct States of Chromatin and Contribute to Polycomb Repression.
Gozalo A, Duke A, Lan Y, Pascual-Garcia P, Talamas JA, Nguyen SC, Shah PP, Jain R, Joyce EF, Capelson M. Gozalo A, et al. Mol Cell. 2020 Jan 2;77(1):67-81.e7. doi: 10.1016/j.molcel.2019.10.017. Epub 2019 Nov 26. Mol Cell. 2020. PMID: 31784359 Free PMC article. - Nucleoporins and transcription: new connections, new questions.
Ikegami K, Lieb JD. Ikegami K, et al. PLoS Genet. 2010 Feb 26;6(2):e1000861. doi: 10.1371/journal.pgen.1000861. PLoS Genet. 2010. PMID: 20195517 Free PMC article. No abstract available. - Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory.
Light WH, Brickner DG, Brand VR, Brickner JH. Light WH, et al. Mol Cell. 2010 Oct 8;40(1):112-25. doi: 10.1016/j.molcel.2010.09.007. Mol Cell. 2010. PMID: 20932479 Free PMC article. - The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway.
Coyle JH, Bor YC, Rekosh D, Hammarskjold ML. Coyle JH, et al. RNA. 2011 Jul;17(7):1344-56. doi: 10.1261/rna.2616111. Epub 2011 May 25. RNA. 2011. PMID: 21613532 Free PMC article.
References
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
- Clapier CR, Cairns BR. The biology of chromatin remodelling complexes. Annu Rev Biochem. 2009;78:273–304. - PubMed
- Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. - PubMed
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous