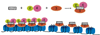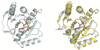Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose - PubMed (original) (raw)
Review
Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose
Lei Tong et al. Biochim Biophys Acta. 2010 Aug.
Abstract
Sirtuins catalyze the NAD(+)-dependent deacetylation of target proteins, which are regulated by this reversible lysine modification. During deacetylation, the glycosidic bond of the nicotinamide ribose is cleaved to yield nicotinamide and the ribose accepts the acetyl group from substrate to produce O-acetyl-ADP-ribose (OAADPr), which exists as an approximately 50:50 mixture of 2' and 3' isomers at neutral pH. Discovery of this metabolite has fueled the idea that OAADPr may play an important role in the biology associated with sirtuins, acting as a signaling molecule and/or an important substrate for downstream enzymatic processes. Evidence for OAADPr-metabolizing enzymes indicates that at least three distinct activities exist that could modulate the cellular levels of this NAD(+)-derived metabolite. In Saccharomyces cerevisiae, NUDIX hydrolase Ysa1 cleaves OAADPr to AMP and 2- and 3-O-acetylribose-5-phosphate, lowering the cellular levels of OAADPr. A buildup of OAADPr and ADPr has been linked to a metabolic shift that lowers endogenous reactive oxygen species and diverts glucose towards preventing oxidative damage. In vitro, the mammalian enzyme ARH3 hydrolyzes OAADPr to acetate and ADPr. A third nuclear-localized activity appears to utilize OAADPr to transfer the acetyl-group to another small molecule, whose identity remains unknown. Recent studies suggest that OAADPr may regulate gene silencing by facilitating the assembly and loading of the Sir2-4 silencing complex onto nucleosomes. In mammalian cells, the Trpm2 cation channel is gated by both OAADPr and ADP-ribose. Binding is mediated by the NUDIX homology (NudT9H) domain found within the intracellular portion of the channel. OAADPr is capable of binding the Macro domain of splice variants from histone protein MacroH2A, which is highly enriched at heterochromatic regions. With recently developed tools, the pace of new discoveries of OAADPr-dependent processes should facilitate new molecular insight into the diverse biological processes modulated by sirtuins.
Copyright 2010 Elsevier B.V. All rights reserved.
Figures
Figure 1. _O_AADPr production by Sir2/sirtuins deacetylation reaction
Sir2 and sirtuins catalyzes NAD+ dependent deacetylation of histone tails, or other non-histone acetylated proteins. The reaction transfers the acetyl group from acetylated lysine residues to the ADP-ribose moiety of NAD+, generating deacetylated histone tails, nicotinamide, and the novel metabolite _O_AADPr.
Figure 2. _O_AADPr metabolism by the ADPr pyrophosphatase, unknown esterase and acetyl transferase
The pyrophosphatase cleaves the pyrophosphate bond of _O_AADPr, generating acetyl-ribose-phosphate and AMP; the esterase removes the acetyl group of _O_AADPr; the unknown acetyl-transferase transfers the acetyl group from _O_AADPr to an unknown molecule X.
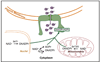
Figure 3. Model for the role of _O_AADPr in facilitating Sir complex assembly and loading onto the chromatin
_O_AADPr interaction with the Sir2-4 complex facilitates loading of the complex onto nucleosomes, probably through binding to Sir2/Sir3. _O_AADPr might bind Sir3 alone and promote Sir3 loading onto nucleosomes.
Figure 4
Left: model of mH2A 1.1 in complex with ADPR. Model generated by overlaying mH2A 1.1 structure (PDB ID: 2FXK) onto a macro domain-ADPR complex (PDB ID: 2BFQ). Two important glycine residues involved in pyrophosphate binding are colored magenta. F348 and D203 side chains are shown in stick representation. Right: the side chain of the Ile in the EIS insertion clashes with the adenine ring. mH2A 1.1 colored cyan, mH2A 1.2 colored yellow.
Figure 5. _O_AADPr and ADPr modulates calcium influx through TRPM2 channel
In mammalian cells, _O_AADPr produced by sirtuins binds the NudT9H domain of TRPM2 and induces calcium influx into the cells. ADPr, another metabolite of NAD+, is believed to gate TRPM2 in a similar mechanism as _O_AADPr.
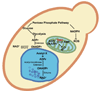
Figure 6. _O_AADPr metabolism and proposed functions in S. cerevisiae
_O_AADPr is produced by sirtuins and can be metabolized by Ysa1, an unknown esterase, and unknown acetyl transferase. Cells lacking Ysa1 display accumulation of ADPr/_O_AADPr and a corresponding decrease in AMP. Increased ADPr/_O_AADPr levels might protect cells against ROS via inhibition of the ETC and generation of higher NADPH levels from increased flux through the pentose phosphate pathway.
Similar articles
- Quantification of endogenous sirtuin metabolite O-acetyl-ADP-ribose.
Lee S, Tong L, Denu JM. Lee S, et al. Anal Biochem. 2008 Dec 15;383(2):174-9. doi: 10.1016/j.ab.2008.08.033. Epub 2008 Sep 7. Anal Biochem. 2008. PMID: 18812159 Free PMC article. - Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3.
Kasamatsu A, Nakao M, Smith BC, Comstock LR, Ono T, Kato J, Denu JM, Moss J. Kasamatsu A, et al. J Biol Chem. 2011 Jun 17;286(24):21110-7. doi: 10.1074/jbc.M111.237636. Epub 2011 Apr 17. J Biol Chem. 2011. PMID: 21498885 Free PMC article. - Analysis of O-acetyl-ADP-ribose as a target for Nudix ADP-ribose hydrolases.
Rafty LA, Schmidt MT, Perraud AL, Scharenberg AM, Denu JM. Rafty LA, et al. J Biol Chem. 2002 Dec 6;277(49):47114-22. doi: 10.1074/jbc.M208997200. Epub 2002 Oct 4. J Biol Chem. 2002. PMID: 12370179 - SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.
Sauve AA, Schramm VL. Sauve AA, et al. Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review. - Mitochondrial sirtuins.
Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Huang JY, et al. Biochim Biophys Acta. 2010 Aug;1804(8):1645-51. doi: 10.1016/j.bbapap.2009.12.021. Epub 2010 Jan 7. Biochim Biophys Acta. 2010. PMID: 20060508 Review.
Cited by
- Sirtuin catalysis and regulation.
Feldman JL, Dittenhafer-Reed KE, Denu JM. Feldman JL, et al. J Biol Chem. 2012 Dec 14;287(51):42419-27. doi: 10.1074/jbc.R112.378877. Epub 2012 Oct 18. J Biol Chem. 2012. PMID: 23086947 Free PMC article. Review. - Biochemistry: Sirtuin on a high-fat diet.
Bheda P, Wolberger C. Bheda P, et al. Nature. 2013 Apr 4;496(7443):41-2. doi: 10.1038/496041a. Nature. 2013. PMID: 23552941 No abstract available. - Proximal ADP-ribose Hydrolysis in Trypanosomatids is Catalyzed by a Macrodomain.
Haikarainen T, Lehtiö L. Haikarainen T, et al. Sci Rep. 2016 Apr 11;6:24213. doi: 10.1038/srep24213. Sci Rep. 2016. PMID: 27064071 Free PMC article. - Interplay between ADP-ribosyltransferases and essential cell signaling pathways controls cellular responses.
Boehi F, Manetsch P, Hottiger MO. Boehi F, et al. Cell Discov. 2021 Nov 2;7(1):104. doi: 10.1038/s41421-021-00323-9. Cell Discov. 2021. PMID: 34725336 Free PMC article. Review. - Emerging characterization of the role of SIRT3-mediated mitochondrial protein deacetylation in the heart.
Sack MN. Sack MN. Am J Physiol Heart Circ Physiol. 2011 Dec;301(6):H2191-7. doi: 10.1152/ajpheart.00199.2011. Epub 2011 Oct 7. Am J Physiol Heart Circ Physiol. 2011. PMID: 21984547 Free PMC article. Review.
References
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. - PubMed
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials