Integrative analysis of the melanoma transcriptome - PubMed (original) (raw)
doi: 10.1101/gr.103697.109. Epub 2010 Feb 23.
Joshua Z Levin, Krishna Vijayendran, Andrey Sivachenko, Xian Adiconis, Jared Maguire, Laura A Johnson, James Robinson, Roel G Verhaak, Carrie Sougnez, Robert C Onofrio, Liuda Ziaugra, Kristian Cibulskis, Elisabeth Laine, Jordi Barretina, Wendy Winckler, David E Fisher, Gad Getz, Matthew Meyerson, David B Jaffe, Stacey B Gabriel, Eric S Lander, Reinhard Dummer, Andreas Gnirke, Chad Nusbaum, Levi A Garraway
Affiliations
- PMID: 20179022
- PMCID: PMC2847744
- DOI: 10.1101/gr.103697.109
Integrative analysis of the melanoma transcriptome
Michael F Berger et al. Genome Res. 2010 Apr.
Abstract
Global studies of transcript structure and abundance in cancer cells enable the systematic discovery of aberrations that contribute to carcinogenesis, including gene fusions, alternative splice isoforms, and somatic mutations. We developed a systematic approach to characterize the spectrum of cancer-associated mRNA alterations through integration of transcriptomic and structural genomic data, and we applied this approach to generate new insights into melanoma biology. Using paired-end massively parallel sequencing of cDNA (RNA-seq) together with analyses of high-resolution chromosomal copy number data, we identified 11 novel melanoma gene fusions produced by underlying genomic rearrangements, as well as 12 novel readthrough transcripts. We mapped these chimeric transcripts to base-pair resolution and traced them to their genomic origins using matched chromosomal copy number information. We also used these data to discover and validate base-pair mutations that accumulated in these melanomas, revealing a surprisingly high rate of somatic mutation and lending support to the notion that point mutations constitute the major driver of melanoma progression. Taken together, these results may indicate new avenues for target discovery in melanoma, while also providing a template for large-scale transcriptome studies across many tumor types.
Figures
Figure 1.
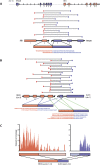
Gene fusions in melanoma. (A) _RB1_-ITM2B gene fusion in short-term culture M990802. ITM2B is transcribed immediately upstream of RB1 on chromosome 13, yet in M990802, a fusion transcript beginning with the 5′ end of RB1 and ending with the 3′ end of ITM2B is implicated by 14 distinct read pairs and two individual fusion-spanning reads. (B) _RECK_-ALX3 gene fusion in short-term culture M000921. Two alternate transcripts are observed in the RNA-seq data: One joining exon 13 of RECK to exon 3 of ALX3, and one joining exon 14 of RECK to exon 3 of ALX3. (C) Spatial distribution of sequence reads for RECK and ALX3 are consistent with a fusion transcript containing the 5′ end of RECK and the 3′ end of ALX3.
Figure 2.
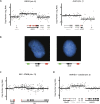
Genomic aberrations leading to gene fusions. (A) Evidence for copy number breakpoints inside RECK and ALX3 from Affymetrix SNP 6.0 microarrays. Raw probe signals were normalized (spots) and segmented (lines) as described in Methods. Exons in red represent those occurring in the _RECK_-ALX3 fusion transcript. (B) FISH in short-term culture M000921 at the RECK and ALX3 loci. Probes positioned 100–200 kb on both sides of each gene reveal amplifications 5′ of RECK and 3′ of ALX3. (C) Amplification involving ITM2B and RB1 on chromosome 13. A tandem duplication would position exon 2 of RB1 upstream of exon 3 of ITM2B (red), as observed in the fusion transcript. (D) Amplification involving C5orf32, ANKHD1, and intervening genes. A tandem duplication would position exon 1 of ANKHD1 upstream of exon 3 of C5orf32 (red), as observed in the fusion transcript.
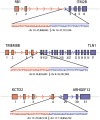
Figure 3.
Genomic breakpoints mapped to base pair resolution. The precise locations of the fusion points in the genomic DNA were determined by Sanger sequencing for three gene fusions: _RB1_-ITM2B, _TMEM8B_-TLN1, and _KCTD2_-ARHGEF12.
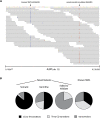
Figure 4.
Co-occurrence of CDKN2A and RB1 deletions. (A) Array CGH copy number profiles for 70 primary cutaneous melanomas (Curtin et al. 2005). Blue segments correspond to deleted regions, and red segments correspond to amplified regions. (B) Deletions at both loci (CDKN2A and RB1) were found to significantly co-occur (P = 0.022).
Figure 5.
Novel recurrent readthrough transcript. (A) _CDK2_-RAB5B, shown here for the MeWo cell line, was independently discovered in four melanomas, with supporting evidence observed in three more. In MeWo, _CDK2_-RAB5B is implicated by eight distinct read pairs and 10 individual junction-spanning reads. (B) Expression level of CDK2 as measured by RNA-seq for 10 melanomas. RPKM, reads per kilobase of exon model per million mapped reads (Mortazavi et al. 2008).
Figure 6.
Validation of somatic mutations. (A) Sequence variants in exon 16 of A2M in melanoma short-term culture M970109. Illumina 51-mer reads are shown as gray boxes (arrowheads denote directionality of reads; nonreference bases are colored and shaded according to their quality scores). One variant (T → C) is homozygous and corresponds to a known SNP in dbSNP (Sherry et al. 2001). The other variant (C → T) is heterozygous and corresponds to a missense E624K mutation. This mutation was validated and confirmed to be somatic. (B) Distribution of transitions and transversions in sequence variants genotyped by Sequenom. Validated somatic mutations were largely CG to TA transitions (86%), representative of mutations induced by UV damage (Drobetsky et al. 1987). In contrast, only 53% of novel germline variants and 0% of variants that failed to validate were CG to TA transitions. Thirty six percent of all known SNPs detected by Illumina in the 10 melanoma samples were CG to TA transitions with respect to human genome reference hg18.
Similar articles
- Genomic and Transcriptomic Analysis Reveals Incremental Disruption of Key Signaling Pathways during Melanoma Evolution.
Shain AH, Joseph NM, Yu R, Benhamida J, Liu S, Prow T, Ruben B, North J, Pincus L, Yeh I, Judson R, Bastian BC. Shain AH, et al. Cancer Cell. 2018 Jul 9;34(1):45-55.e4. doi: 10.1016/j.ccell.2018.06.005. Cancer Cell. 2018. PMID: 29990500 Free PMC article. - A Transcriptome-Wide Isoform Landscape of Melanocytic Nevi and Primary Melanomas Identifies Gene Isoforms Associated with Malignancy.
Hakobyan S, Loeffler-Wirth H, Arakelyan A, Binder H, Kunz M. Hakobyan S, et al. Int J Mol Sci. 2021 Jul 2;22(13):7165. doi: 10.3390/ijms22137165. Int J Mol Sci. 2021. PMID: 34281234 Free PMC article. - Integrative genome comparison of primary and metastatic melanomas.
Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C, Granter SR, Ramaswamy S, Golub T, Duncan LM, Wagner SN, Brennan C, Chin L. Kabbarah O, et al. PLoS One. 2010 May 24;5(5):e10770. doi: 10.1371/journal.pone.0010770. PLoS One. 2010. PMID: 20520718 Free PMC article. - Applications of genomics in melanoma oncogene discovery.
Berger MF, Garraway LA. Berger MF, et al. Hematol Oncol Clin North Am. 2009 Jun;23(3):397-414, vii. doi: 10.1016/j.hoc.2009.03.005. Hematol Oncol Clin North Am. 2009. PMID: 19464593 Free PMC article. Review. - Melanocytic Skin Neoplasms: What Lesson From Genomic Aberrations?
Urso C. Urso C. Am J Dermatopathol. 2019 Sep;41(9):623-629. doi: 10.1097/DAD.0000000000001341. Am J Dermatopathol. 2019. PMID: 31433323 Review.
Cited by
- Visualizing Subcellular Enrichment of Glycogen in Live Cancer Cells by Stimulated Raman Scattering.
Lee D, Du J, Yu R, Su Y, Heath JR, Wei L. Lee D, et al. Anal Chem. 2020 Oct 6;92(19):13182-13191. doi: 10.1021/acs.analchem.0c02348. Epub 2020 Sep 21. Anal Chem. 2020. PMID: 32907318 Free PMC article. - Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer.
Wu C, Wyatt AW, Lapuk AV, McPherson A, McConeghy BJ, Bell RH, Anderson S, Haegert A, Brahmbhatt S, Shukin R, Mo F, Li E, Fazli L, Hurtado-Coll A, Jones EC, Butterfield YS, Hach F, Hormozdiari F, Hajirasouliha I, Boutros PC, Bristow RG, Jones SJ, Hirst M, Marra MA, Maher CA, Chinnaiyan AM, Sahinalp SC, Gleave ME, Volik SV, Collins CC. Wu C, et al. J Pathol. 2012 May;227(1):53-61. doi: 10.1002/path.3987. Epub 2012 Mar 21. J Pathol. 2012. PMID: 22294438 Free PMC article. - Whole genome sequencing of matched primary and metastatic acral melanomas.
Turajlic S, Furney SJ, Lambros MB, Mitsopoulos C, Kozarewa I, Geyer FC, Mackay A, Hakas J, Zvelebil M, Lord CJ, Ashworth A, Thomas M, Stamp G, Larkin J, Reis-Filho JS, Marais R. Turajlic S, et al. Genome Res. 2012 Feb;22(2):196-207. doi: 10.1101/gr.125591.111. Epub 2011 Dec 19. Genome Res. 2012. PMID: 22183965 Free PMC article. - Highlights from the prostate cancer genome report.
Tan SH, Petrovics G, Srivastava S. Tan SH, et al. Asian J Androl. 2011 Sep;13(5):659-60. doi: 10.1038/aja.2011.84. Epub 2011 Jun 13. Asian J Androl. 2011. PMID: 21666698 Free PMC article. No abstract available. - Integrative network analysis identifies key genes and pathways in the progression of hepatitis C virus induced hepatocellular carcinoma.
Zheng S, Tansey WP, Hiebert SW, Zhao Z. Zheng S, et al. BMC Med Genomics. 2011 Aug 8;4:62. doi: 10.1186/1755-8794-4-62. BMC Med Genomics. 2011. PMID: 21824427 Free PMC article.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. - PubMed
- Blacker D, Wilcox MA, Laird NM, Rodes L, Horvath SM, Go RC, Perry R, Watson B, Jr, Bassett SS, McInnis MG, et al. Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet. 1998;19:357–360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical