Distinguishing epigenetic marks of developmental and imprinting regulation - PubMed (original) (raw)
Distinguishing epigenetic marks of developmental and imprinting regulation
Kirsten R McEwen et al. Epigenetics Chromatin. 2010.
Abstract
Background: The field of epigenetics is developing rapidly, however we are only beginning to comprehend the complexity of its influence on gene regulation. Using genomic imprinting as a model we examine epigenetic profiles associated with different forms of gene regulation. Imprinting refers to the expression of a gene from only one of the chromosome homologues in a parental-origin-specific manner. This is dependent on heritable germline epigenetic control at a cis-acting imprinting control region that influences local epigenetic states. Epigenetic modifications associated with imprinting regulation can be compared to those associated with the more canonical developmental regulation, important for processes such as differentiation and tissue specificity. Here we test the hypothesis that these two mechanisms are associated with different histone modification enrichment patterns.
Results: Using high-throughput data extraction with subsequent analysis, we have found that particular histone modifications are more likely to be associated with either imprinting repression or developmental repression of imprinted genes. H3K9me3 and H4K20me3 are together enriched at imprinted genes with differentially methylated promoters and do not show a correlation with developmental regulation. H3K27me3 and H3K4me3, however, are more often associated with developmental regulation. We find that imprinted genes are subject to developmental regulation through bivalency with H3K4me3 and H3K27me3 enrichment on the same allele. Furthermore, a specific tri-mark signature comprising H3K4me3, H3K9me3 and H4K20me3 has been identified at all imprinting control regions.
Conclusion: A large amount of data is produced from whole-genome expression and epigenetic profiling studies of cellular material. We have shown that such publicly available data can be mined and analysed in order to generate novel findings for categories of genes or regulatory elements. Comparing two types of gene regulation, imprinting and developmental, our results suggest that different histone modifications associate with these distinct processes. This form of analysis is therefore a useful tool to elucidate the complex epigenetic code associated with genome function and to determine the underlying features conferring epigenetic states.
Figures
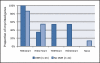
Figure 1
Histone modification patterns in mouse embryonic stem cells (ESCs). Profiles for H3K4me3, H3K27me3 and H3K9me3 were analysed in mouse ESCs at transcription start sites of (A) imprinted genes (n = 54) and (B) all genes (n = 17,761) using source enrichment data of Mikkelsen et al. [50]. 'Other' represents alternative combinations of these three histone modifications and 'None' represents genes without any of these three modifications. The data does not meet the conditions of the chi-square test when comparing all profiles and also when specifically comparing H3K4me3 and H3K9me3 between imprinted and all genes. H3K4me3 and H3K27me3 are together enriched more often at imprinted genes than all genes (Yates' chi-square test, P < 0.005).
Figure 2
Impact of promoter differential methylation on histone modification profiles at imprinted genes. Histone modification enrichment is compared in mouse embryonic stem cells at imprinted genes with and without a promoter differentially methylated region (DMR). Transcription start sites were assessed for enrichment of H3K4me3, H3K27me3, H3K9me3 and H4K20me3 using source data from Mikkelsen et al. [50]. The presence of one particular modification at an imprinted gene does not preclude the presence of another. A significantly different epigenetic profile for these four marks is observed at genes with a promoter DMR compared to genes without a promoter DMR (chi-square contingency test, P < 0.0001). Germline DMRs are not distinguished from somatic DMRs in this analysis. H3K9me3 and H4K20me3 are exclusively enriched at imprinted genes possessing a promoter DMR. A greater proportion of genes with promoter DMRs are developmentally expressed compared to genes without promoter DMRs (71% compared to 48% respectively); the increase in H3K4me3 and the decrease in H3K27me3 at genes with a promoter DMR likely reflects this (see Additional file 2 and Figure 4).
Figure 3
Histone modification patterns at imprinted genes across cell types. Histone modification profiles for H3K4me3 and H3K27me3 were analysed at transcription start sites of imprinted genes (n = 54) in mouse (A) embryonic stem cells (ESCs), (B) neural progenitor cells (NPCs) and (C) mouse embryonic fibroblasts (MEFs) using Mikkelsen et al. [50] whole-genome source data. The patterns of enrichment shown differ significantly across the three cell types (chi-square contingency test, P < 0.0005). Coenrichment of H3K4me3 and H3K27me3 is significantly higher at ESCs compared to NPCs (Yates' chi-square test, P < 0.01). A reduction from 41% in ESCs to 22% in MEFs is observed for this specific profile (Yates' chi-square test, P = 0.123).
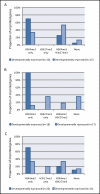
Figure 4
Enrichment of H3K4me3 and H3K27me3 with respect to developmental expression status. The two histone modifications H3K4me3 and H3K27me3 are shown to be important for the developmental regulation of imprinted genes. Using Mikkensen et al. [50] as a source for histone modification enrichment and expression data, histone modification profiles of H3K4me3 and H3K27me3 at imprinted gene transcription start sites were assessed in mouse (A) embryonic stem cells (ESCs), (B) neural progenitor cells (NPCs) and (C) mouse embryonic fibroblasts (MEFs) at developmentally expressed and repressed imprinted genes. Though a trend is observed, profiles at developmentally expressed imprinted genes do not differ significantly to repressed genes for ESCs or MEFs (chi-square contingency test, ESC:P = 0.097, MEF: P = 0.079). NPC data does not meet the conditions required for the chi-square contingency test.
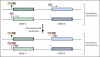
Figure 5
Working model of histone modification profiles at imprinted genes undergoing developmental repression. In mouse embryonic stem cells, the presence of a germline differentially methylated region reflecting an imprinting control region at the promoter of gene A confers a distinct histone modification profile to that of gene B, which has no differential DNA methylation; gene A is marked by H3K4me3 on the active allele and by H3K9me3 plus H4K20me3 on the inactive allele. After developmental repression, both genes acquire H3K27me3 on the previously active allele. Transcriptional activators and repressors are key players in this process and may be a cause or consequence of the histone modification states shown. All histone modifications illustrated represent the trimethylated state.
Similar articles
- Cross-species clues of an epigenetic imprinting regulatory code for the IGF2R gene.
Vu TH, Jirtle RL, Hoffman AR. Vu TH, et al. Cytogenet Genome Res. 2006;113(1-4):202-8. doi: 10.1159/000090833. Cytogenet Genome Res. 2006. PMID: 16575181 Review. - Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions.
Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL. Dindot SV, et al. Genome Res. 2009 Aug;19(8):1374-83. doi: 10.1101/gr.089185.108. Epub 2009 Jun 19. Genome Res. 2009. PMID: 19542493 Free PMC article. - [Epigenetics, genomic imprinting and developmental disorders].
Le Bouc Y, Rossignol S, Azzi S, Brioude F, Cabrol S, Gicquel C, Netchine I. Le Bouc Y, et al. Bull Acad Natl Med. 2010 Feb;194(2):287-97; discussion 297-300. Bull Acad Natl Med. 2010. PMID: 21166119 French. - Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues.
Hanna CW, Pérez-Palacios R, Gahurova L, Schubert M, Krueger F, Biggins L, Andrews S, Colomé-Tatché M, Bourc'his D, Dean W, Kelsey G. Hanna CW, et al. Genome Biol. 2019 Oct 29;20(1):225. doi: 10.1186/s13059-019-1833-x. Genome Biol. 2019. PMID: 31665063 Free PMC article. - Canonical and Non-canonical Genomic Imprinting in Rodents.
Kobayashi H. Kobayashi H. Front Cell Dev Biol. 2021 Aug 5;9:713878. doi: 10.3389/fcell.2021.713878. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34422832 Free PMC article. Review.
Cited by
- The Epigenetics of Normal Pregnancy.
Best JD, Carey N. Best JD, et al. Obstet Med. 2013 Mar;6(1):3-7. doi: 10.1258/OM.2011.110070. Epub 2013 Mar 1. Obstet Med. 2013. PMID: 27757144 Free PMC article. Review. - Homeobox gene Rhox5 is regulated by epigenetic mechanisms in cancer and stem cells and promotes cancer growth.
Li Q, O'Malley ME, Bartlett DL, Guo ZS. Li Q, et al. Mol Cancer. 2011 May 24;10:63. doi: 10.1186/1476-4598-10-63. Mol Cancer. 2011. PMID: 21609483 Free PMC article. - A Decade of Exploring the Mammalian Sperm Epigenome: Paternal Epigenetic and Transgenerational Inheritance.
Champroux A, Cocquet J, Henry-Berger J, Drevet JR, Kocer A. Champroux A, et al. Front Cell Dev Biol. 2018 May 15;6:50. doi: 10.3389/fcell.2018.00050. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 29868581 Free PMC article. Review. - Methylome Analysis in Chickens Immunized with Infectious Laryngotracheitis Vaccine.
Carrillo JA, He Y, Luo J, Menendez KR, Tablante NL, Zhao K, Paulson JN, Li B, Song J. Carrillo JA, et al. PLoS One. 2015 Jun 24;10(6):e0100476. doi: 10.1371/journal.pone.0100476. eCollection 2015. PLoS One. 2015. PMID: 26107953 Free PMC article. - Transgene- and locus-dependent imprinting reveals allele-specific chromosome conformations.
Lonfat N, Montavon T, Jebb D, Tschopp P, Nguyen Huynh TH, Zakany J, Duboule D. Lonfat N, et al. Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):11946-51. doi: 10.1073/pnas.1310704110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818637 Free PMC article.