Molecular basis of the core structure of tight junctions - PubMed (original) (raw)
Review
Molecular basis of the core structure of tight junctions
Mikio Furuse. Cold Spring Harb Perspect Biol. 2010 Jan.
Abstract
The morphological feature of tight junctions (TJs) fits well with their functions. The core of TJs is a fibril-like proteinaceous structure within the lipid bilayer, the so-called TJ strands. TJ strands in apposing plasma membranes associate with each other to eliminate the intercellular space. A network of paired TJ strands generates a continuous belt that circumscribes each cell to establish the diffusion barrier to the solutes in the paracellular pathway throughout the cellular sheet. Identification and characterization of TJ-associated proteins during the last two decades has unveiled the nature of TJ strands and how they are spatially organized. The interplay between integral membrane proteins, claudins, and cytoplasmic plaque proteins, ZO-1/ZO-2, is critical for TJ formation and function.
Figures
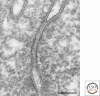
Figure 1.
Thin section of TJs of mouse epididymis epithelial cells. TJs are defined as close contacts between plasma membranes of adjacent cells. Bar, 100 nm.
Figure 2.
Freeze-fracture replica of TJs of mouse intestinal epithelial cells. TJs appear as anastomosing fibrils, namely, TJ strands, on P-face (arrow) or complementary grooves on E-face (arrowhead) in glutaraldehyde-fixed specimens. Bar, 200 nm.
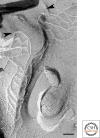
Figure 3.
Freeze-fracture replica of tTJs in MDCK cells. At tricellular corners, the most apical elements of the horizontal TJ strands from both sides (arrowheads) attach to form the central seal and extend vertically in the basal direction. Short, horizontal strands connect with the central seal. Bar, 100 nm.
Figure 4.
The membrane-spanning model of claudins and the domain organization of claudin-binding plaque proteins. Claudins contain four transmembrane domains. The conserved amino acid residues in the first extracellular loop are indicated. The PDZ domain binding motif is located at the carboxyl terminus, which is known to bind to PDZ-domains in ZO-1, ZO-2, ZO-3, MUPP1, and PATJ (stars). (GUK) guanylate kinase domain; (L27) LIN2, 7 homology domain; (U5) unique 5 region; (U6) unique 6 region. U6 contains the acidic region.
Similar articles
- Claudins and JAM-A coordinately regulate tight junction formation and epithelial polarity.
Otani T, Nguyen TP, Tokuda S, Sugihara K, Sugawara T, Furuse K, Miura T, Ebnet K, Furuse M. Otani T, et al. J Cell Biol. 2019 Oct 7;218(10):3372-3396. doi: 10.1083/jcb.201812157. Epub 2019 Aug 29. J Cell Biol. 2019. PMID: 31467165 Free PMC article. - Claudin-7 expressed on lateral membrane of rat epididymal epithelium does not form aberrant tight junction strands.
Inai T, Sengoku A, Hirose E, Iida H, Shibata Y. Inai T, et al. Anat Rec (Hoboken). 2007 Nov;290(11):1431-8. doi: 10.1002/ar.20597. Anat Rec (Hoboken). 2007. PMID: 17853415 - The Claudins: From Tight Junctions to Biological Systems.
Tsukita S, Tanaka H, Tamura A. Tsukita S, et al. Trends Biochem Sci. 2019 Feb;44(2):141-152. doi: 10.1016/j.tibs.2018.09.008. Epub 2018 Oct 25. Trends Biochem Sci. 2019. PMID: 30665499 Review. - Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier.
Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Sonoda N, et al. J Cell Biol. 1999 Oct 4;147(1):195-204. doi: 10.1083/jcb.147.1.195. J Cell Biol. 1999. PMID: 10508866 Free PMC article. - Tight junction-based epithelial microenvironment and cell proliferation.
Tsukita S, Yamazaki Y, Katsuno T, Tamura A, Tsukita S. Tsukita S, et al. Oncogene. 2008 Nov 24;27(55):6930-8. doi: 10.1038/onc.2008.344. Oncogene. 2008. PMID: 19029935 Review.
Cited by
- The endothelium permeability after bioresorbable scaffolds implantation caused by the heterogeneous expression of tight junction proteins.
Huang J, Ge S, Luo D, Du R, Wang Y, Liu W, Wang G, Yin T. Huang J, et al. Mater Today Bio. 2022 Aug 27;16:100410. doi: 10.1016/j.mtbio.2022.100410. eCollection 2022 Dec. Mater Today Bio. 2022. PMID: 36090609 Free PMC article. - Blood cells and endothelial barrier function.
Rodrigues SF, Granger DN. Rodrigues SF, et al. Tissue Barriers. 2015 Apr 3;3(1-2):e978720. doi: 10.4161/21688370.2014.978720. eCollection 2015. Tissue Barriers. 2015. PMID: 25838983 Free PMC article. - Claudin-4 is required for vasculogenic mimicry formation in human breast cancer cells.
Cui YF, Liu AH, An DZ, Sun RB, Shi Y, Shi YX, Shi M, Zhang Q, Wang LL, Feng Q, Pan GL, Wang Q. Cui YF, et al. Oncotarget. 2015 May 10;6(13):11087-97. doi: 10.18632/oncotarget.3571. Oncotarget. 2015. PMID: 25871476 Free PMC article. - Tight junctions in lung cancer and lung metastasis: a review.
Soini Y. Soini Y. Int J Clin Exp Pathol. 2012;5(2):126-36. Epub 2012 Feb 12. Int J Clin Exp Pathol. 2012. PMID: 22400072 Free PMC article. Review. - The functional antagonist of sphingosine-1-phosphate, FTY720, impairs gut barrier function.
Sikdar S, Mitra D, Das O, Bhaumik M, Dutta S. Sikdar S, et al. Front Pharmacol. 2024 Aug 19;15:1407228. doi: 10.3389/fphar.2024.1407228. eCollection 2024. Front Pharmacol. 2024. PMID: 39224783 Free PMC article.
References
- Anderson JM, Cereijido M 2001. Evolution of ideas on the tight junction. In Tight junctions, 2nd ed. (ed. Cereijido M, Anderson J), pp. 1–18 CRC Press, Boca Raton
- Aono S, Hirai Y 2008. Phosphorylation of claudin-4 is required for tight junction formation in a human keratinocyte cell line. Exp Cell Res 314:3326–3339 - PubMed
- Balda MS, Anderson JM 1993. Two classes of tight junctions are revealed by ZO-1 isoforms. Am J Physiol 264:918–924 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources