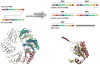Bacterial microcompartment organelles: protein shell structure and evolution - PubMed (original) (raw)
Review
Bacterial microcompartment organelles: protein shell structure and evolution
Todd O Yeates et al. Annu Rev Biophys. 2010.
Abstract
Some bacteria contain organelles or microcompartments consisting of a large virion-like protein shell encapsulating sequentially acting enzymes. These organized microcompartments serve to enhance or protect key metabolic pathways inside the cell. The variety of bacterial microcompartments provide diverse metabolic functions, ranging from CO(2) fixation to the degradation of small organic molecules. Yet they share an evolutionarily related shell, which is defined by a conserved protein domain that is widely distributed across the bacterial kingdom. Structural studies on a number of these bacterial microcompartment shell proteins are illuminating the architecture of the shell and highlighting its critical role in controlling molecular transport into and out of microcompartments. Current structural, evolutionary, and mechanistic ideas are discussed, along with genomic studies for exploring the function and diversity of this family of bacterial organelles.
Figures
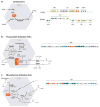
Figure 1. Electron micrographs of various bacterial microcompartments
(a) Transmission electron micrographs showing (left) a section through a dividing cyanobacterial cell (Synechocystis sp. PCC 6803) and (right) an enlargement of a single carboxysome on the right (courtesy of Wim Vermaas) (adapted from Reference 39). (b) Purified carboxysomes from Halothiobacillus neapolitanus (sample courtesy of Sabine Heinhorst and Gordon Cannon, image courtesy of Kelly Dryden and Mark Yeager). (c) Isolated Pdu microcompartments from Salmonella enterica serovar Typhimurium LT2 (courtesy of Thomas Bobik).
Figure 2
Gene organization and proposed metabolic pathways for three types of bacterial microcompartments. Genes are colored to indicate their homology. All BMC shell proteins are light blue. For each microcompartment, the key sequestered intermediate is boxed in orange. (a) Function of the carboxysome in enhancing CO2 fixation. See text and Reference for mechanistic details. Gene organizations for α- and β-carboxysomes are on the right. (b) A current model for the function of the propanediol utilization (Pdu) microcompartment in metabolizing 1,2-propanediol. See text and Reference for mechanistic details. The gene organization for the pdu operon is shown on the right. (c) A hypothetical model for the metabolism of ethanolamine in the Eut microcompartment. See text and Reference for mechanistic details. The gene organization of the eut operon is shown on the right.
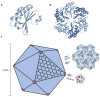
Figure 3
Idealized model for assembly of the carboxysome and related bacterial microcompartments. (a) A ribbon diagram of a typical bacterial microcompartment (BMC) shell protein fold. (b) A hexameric assembly of a BMC protein in a ribbon diagram. (c) Hexameric building blocks of the BMC proteins can assemble into a molecular layer (right), which forms flat facets of the polyhedral shells of various bacterial microcompartments. The pentameric proteins (CcmL or CsoS4A) from the carboxysome (bottom, right) have been argued to form vertices of the icosahedral carboxysome (left) (68). The Pdu and Eut microcompartments are less geometrically regular than the carboxysome and are potentially more complex.
Figure 4
Sequence conservation among diverse bacterial microcompartment (BMC) shell proteins. Conserved amino acid positions (red) were defined as those having sequence identity above 80% in an alignment of 2174 BMC sequences. Positions of high conservation occur mainly at the perimeter, where hexamers meet. In the CcmK1 protein, these residues include A4, G6, A19, D21, K25, V29, G38, G48, V50, V53, and G70; the conserved residue in the pore is glycine G38.
Figure 5
Variations on the bacterial microcompartment (BMC) protein fold. (Top) Secondary structure schematics of BMC proteins in their various arrangements. Individual secondary structure elements are colored. The canonical BMCs include CcmK1/2/3/4, CsoS1A/B/C, PduA/J, and EutM. CcmO likely encodes tandem canonical BMC domains. The permuted, single-domain BMC proteins include PduU and EutS. (Bottom, right) The cores of canonical and permuted BMC proteins are in close agreement, as shown by a superposition of PduU (salmon) over CcmK2 (yellow). (Bottom, left) Both EutL and CsoS1D have permuted BMC domains in tandem, but their tertiary arrangements differ. Individual BMC domains are colored separately. When the N-terminal BMC domains (blue) of the two proteins are superimposed, their C-terminal domains (CsoS1D in magenta and EutL in green) adopt different positions in the hexamer. The linker regions between domains are colored yellow.
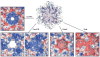
Figure 6
Various pores of bacterial microcompartment (BMC) proteins. Top views of central pores from some representative BMC proteins colored by electrostatic potential (positive: blue; negative: red). Canonical BMC proteins (e.g., CcmK1 from the β-carboxysome) have a small pore at the sixfold, which has a diameter of 4 to 6 Å (39, 69, 72). A circularly permutated tandem BMC protein from the α-carboxysome, CsoS1D, adopts alternative conformations with open and closed pores (40). A circularly permutated BMC protein from the pdu microcompartment, PduU, revealed a totally closed pore (19). Whether this pore opens for transport is unknown. Another circularly permutated tandem BMC protein, EutL, has been observed in two distinct conformations, open (70) (not shown) and closed (40, 70) (shown). See Supplemental Figure 2 for additional views of the pores.
Figure 7
Schematic for identifying conserved bacterial microcompartment (BMC)-proximal protein families. First, BMC homologues were retrieved from the NCBI database (CDD ID cl01982). Their respective chromosomal positions were located in fully sequenced bacterial genomes deposited into the EBI Integr8 database. For each chromosomal position identified, 10 BMC-proximal open reading frames in both the 5′ and 3′ direction were retrieved as candidate microcompartment-associated genes and subsequently assigned to homologous sequence clusters by performing a full pairwise BLAST analysis. The dominant clusters suggest likely microcompartment-associated functions (Supplemental Tables 2 and 3).
Similar articles
- The protein shells of bacterial microcompartment organelles.
Yeates TO, Thompson MC, Bobik TA. Yeates TO, et al. Curr Opin Struct Biol. 2011 Apr;21(2):223-31. doi: 10.1016/j.sbi.2011.01.006. Curr Opin Struct Biol. 2011. PMID: 21315581 Free PMC article. Review. - Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria.
Jorda J, Lopez D, Wheatley NM, Yeates TO. Jorda J, et al. Protein Sci. 2013 Feb;22(2):179-95. doi: 10.1002/pro.2196. Epub 2013 Jan 4. Protein Sci. 2013. PMID: 23188745 Free PMC article. - Polyhedral organelles compartmenting bacterial metabolic processes.
Bobik TA. Bobik TA. Appl Microbiol Biotechnol. 2006 May;70(5):517-25. doi: 10.1007/s00253-005-0295-0. Epub 2006 Mar 9. Appl Microbiol Biotechnol. 2006. PMID: 16525780 Review. - Bacterial microcompartments: their properties and paradoxes.
Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Cheng S, et al. Bioessays. 2008 Nov;30(11-12):1084-95. doi: 10.1002/bies.20830. Bioessays. 2008. PMID: 18937343 Free PMC article. Review. - Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell.
Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. Crowley CS, et al. J Biol Chem. 2010 Nov 26;285(48):37838-46. doi: 10.1074/jbc.M110.160580. Epub 2010 Sep 24. J Biol Chem. 2010. PMID: 20870711 Free PMC article.
Cited by
- MCPdb: The bacterial microcompartment database.
Ochoa JM, Bair K, Holton T, Bobik TA, Yeates TO. Ochoa JM, et al. PLoS One. 2021 Mar 29;16(3):e0248269. doi: 10.1371/journal.pone.0248269. eCollection 2021. PLoS One. 2021. PMID: 33780471 Free PMC article. - Prokaryotic gene clusters: a rich toolbox for synthetic biology.
Fischbach M, Voigt CA. Fischbach M, et al. Biotechnol J. 2010 Dec;5(12):1277-96. doi: 10.1002/biot.201000181. Biotechnol J. 2010. PMID: 21154668 Free PMC article. Review. - Exploring bacterial organelle interactomes: a model of the protein-protein interaction network in the Pdu microcompartment.
Jorda J, Liu Y, Bobik TA, Yeates TO. Jorda J, et al. PLoS Comput Biol. 2015 Feb 3;11(2):e1004067. doi: 10.1371/journal.pcbi.1004067. eCollection 2015 Feb. PLoS Comput Biol. 2015. PMID: 25646976 Free PMC article. - Genome Sequence of the Autotrophic Acetogen Clostridium magnum DSM 2767.
Uhlig R, Poehlein A, Fischer RJ, Daniel R, Bahl H. Uhlig R, et al. Genome Announc. 2016 Jun 9;4(3):e00464-16. doi: 10.1128/genomeA.00464-16. Genome Announc. 2016. PMID: 27284147 Free PMC article. - Engineered biological entities for drug delivery and gene therapy protein nanoparticles.
Domingo-Espín J, Unzueta U, Saccardo P, Rodríguez-Carmona E, Corchero JL, Vázquez E, Ferrer-Miralles N. Domingo-Espín J, et al. Prog Mol Biol Transl Sci. 2011;104:247-98. doi: 10.1016/B978-0-12-416020-0.00006-1. Prog Mol Biol Transl Sci. 2011. PMID: 22093221 Free PMC article. Review.
References
- Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–22. - PubMed
- Baker SH, Williams DS, Aldrich HC, Gambrell AC, Shively JM. Identification and localization of the carboxysome peptide Csos3 and its corresponding gene in Thiobacillus neapolitanus. Arch Microbiol. 2000;173:278–83. - PubMed
- Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 GM031299/GM/NIGMS NIH HHS/United States
- P01 GM031299-23/GM/NIGMS NIH HHS/United States
- R01 AI081146/AI/NIAID NIH HHS/United States
- AI081146/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources