The large-scale organization of the bacterial network of ecological co-occurrence interactions - PubMed (original) (raw)
The large-scale organization of the bacterial network of ecological co-occurrence interactions
Shiri Freilich et al. Nucleic Acids Res. 2010 Jul.
Abstract
In their natural environments, microorganisms form complex systems of interactions. Understating the structure and organization of bacterial communities is likely to have broad medical and ecological consequences, yet a comprehensive description of the network of environmental interactions is currently lacking. Here, we mine co-occurrences in the scientific literature to construct such a network and demonstrate an expected pattern of association between the species' lifestyle and the recorded number of co-occurring partners. We further focus on the well-annotated gut community and show that most co-occurrence interactions of typical gut bacteria occur within this community. The network is then clustered into species-groups that significantly correspond with natural occurring communities. The relationships between resource competition, metabolic yield and growth rate within the clusters correspond with the r/K selection theory. Overall, these results support the constructed clusters as a first approximation of a bacterial ecosystem model. This comprehensive collection of predicted communities forms a new data resource for further systematic characterization of the ecological design principals shaping communities. Here, we demonstrate its utility for predicting cooperation and inhibition within communities.
Figures
Figure 1.
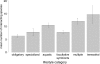
Mean number of co-occurring partners identified for species of different lifestyle. Annotations of lifestyle are according to (25), lifestyle categories are ordered according to their complexity from the most simple communities of obligatory host-associated species to the most complex terrestrial communities (‘Materials and Methods’ section). Number of species in each category are (ordered as in the figure): 27, 5, 4, 42, 28, 3. Bars show the standard error.
Figure 2.
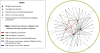
Co-occurrence interactions between obligatory gut bacteria and other bacterial species. Typical gut bacteria (B. fragilis, B. adolescentis, B. longum, B. thetaiotaomicron and _E. faecalis_—represented by black dots) interact with 41 species. Manual survey of these species identified 28 of them as facultative gut bacteria (represented by red dots), five as non-gut human-associated bacteria (green dots) and eight as non-human associated species (blue dots). Overall, the five typical gut bacteria form 69 co-occurrence interactions, 54 of them (78%) are among themselves (six co-occurrence interactions, red lines) and with facultative gut species (48 co-occurrence interactions, black lines). Only 15 co-occurrence interactions are with non-gut species (red, purple and blue lines). The full list of co-occurrence interactions, species, and ecological annotations is provided at Text S1 in the
Supplementary Data
.
Figure 3.
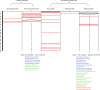
Correspondence between unsupervised, literature-driven clusters and ecological groups, delineating three clusters which are enriched in specialized bacteria, eight in host-associated bacteria, 13 in gut bacteria, two in bacteria from sludge samples and six in marine bacteria. Rows denote clusters (ordered according to their size), columns denote ecological groups. Enriched clusters (detected as described at the ‘Materials and Methods’ section) are shown in red. Only ecological groups (lifestyle categories and environmental sample types) for which at least a single cluster is enriched are shown (data is provided in
Supplementary Table S5
in the
Supplementary Data
). The full list of species classified to cluster 11 and 47 discussed in the main text is detailed in the figure. Species in blue are classified into a specific ecological group (marine species according to detection of species in environmental sample; oral species according to manually curated list composed of five typical oral species). The remaining species within these clusters were manually examined for their ecological distribution; species colored in green are those that can inhabit the corresponding ecological cluster (Text S1 in the
Supplementary Data
), species colored in red are those whose ecological habitat is different.
Figure 4.
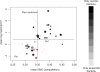
Level of metabolic competition versus growth rate in 38 clusters where growth rate information is available for a least half of the cluster members. Sizes of dots correspond to the sizes of the clusters. Colors of dots correspond to the anaerobic/aerobic ratio. Red dots correspond to cluster which do not contain any aerobic or anaerobic bacteria (alternative annotations include facultative, microaerophilic and unknown species). The plot is divided according to median values of the axes. The clusters indicated by arrows are described at Text S1 in the
Supplementary Data
; the ecological attributes of all clusters are provided in
Table S6
in the
Supplementary Data
. Abbreviations: EMO—effective metabolic overlap; DT—duplication time.
Similar articles
- Bacterial community dynamics as a result of growth-yield trade-off and multispecies metabolic interactions toward understanding the gut biofilm niche.
Valiei A, Dickson AM, Aminian-Dehkordi J, Mofrad MRK. Valiei A, et al. BMC Microbiol. 2024 Oct 29;24(1):441. doi: 10.1186/s12866-024-03566-0. BMC Microbiol. 2024. PMID: 39472801 Free PMC article. - Contrasting Network Features between Free-Living and Particle-Attached Bacterial Communities in Taihu Lake.
Xu H, Zhao D, Huang R, Cao X, Zeng J, Yu Z, Hooker KV, Hambright KD, Wu QL. Xu H, et al. Microb Ecol. 2018 Aug;76(2):303-313. doi: 10.1007/s00248-017-1131-7. Epub 2018 Jan 9. Microb Ecol. 2018. PMID: 29318328 - Competitive and cooperative metabolic interactions in bacterial communities.
Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. Freilich S, et al. Nat Commun. 2011 Dec 13;2:589. doi: 10.1038/ncomms1597. Nat Commun. 2011. PMID: 22158444 - Multispecies Swarms of Social Microorganisms as Moving Ecosystems.
Ben-Jacob E, Finkelshtein A, Ariel G, Ingham C. Ben-Jacob E, et al. Trends Microbiol. 2016 Apr;24(4):257-269. doi: 10.1016/j.tim.2015.12.008. Epub 2016 Jan 25. Trends Microbiol. 2016. PMID: 26822253 Review. - Microbial interactions and community assembly at microscales.
Cordero OX, Datta MS. Cordero OX, et al. Curr Opin Microbiol. 2016 Jun;31:227-234. doi: 10.1016/j.mib.2016.03.015. Epub 2016 May 25. Curr Opin Microbiol. 2016. PMID: 27232202 Free PMC article. Review.
Cited by
- Towards a predictive systems-level model of the human microbiome: progress, challenges, and opportunities.
Greenblum S, Chiu HC, Levy R, Carr R, Borenstein E. Greenblum S, et al. Curr Opin Biotechnol. 2013 Aug;24(4):810-20. doi: 10.1016/j.copbio.2013.04.001. Epub 2013 Apr 23. Curr Opin Biotechnol. 2013. PMID: 23623295 Free PMC article. Review. - NetCooperate: a network-based tool for inferring host-microbe and microbe-microbe cooperation.
Levy R, Carr R, Kreimer A, Freilich S, Borenstein E. Levy R, et al. BMC Bioinformatics. 2015 May 17;16(1):164. doi: 10.1186/s12859-015-0588-y. BMC Bioinformatics. 2015. PMID: 25980407 Free PMC article. - NetMet: A Network-Based Tool for Predicting Metabolic Capacities of Microbial Species and their Interactions.
Tal O, Selvaraj G, Medina S, Ofaim S, Freilich S. Tal O, et al. Microorganisms. 2020 Jun 3;8(6):840. doi: 10.3390/microorganisms8060840. Microorganisms. 2020. PMID: 32503277 Free PMC article. - Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies.
Shetty SA, Hugenholtz F, Lahti L, Smidt H, de Vos WM. Shetty SA, et al. FEMS Microbiol Rev. 2017 Mar 1;41(2):182-199. doi: 10.1093/femsre/fuw045. FEMS Microbiol Rev. 2017. PMID: 28364729 Free PMC article. Review. - Informatics technology mimics ecology: dense, mutualistic collaboration networks are associated with higher publication rates.
Sorani MD. Sorani MD. PLoS One. 2012;7(1):e30463. doi: 10.1371/journal.pone.0030463. Epub 2012 Jan 18. PLoS One. 2012. PMID: 22279593 Free PMC article.
References
- Losos JB, Leal M, Glor RE, De Queiroz K, Hertz PE, Rodriguez Schettino L, Lara AC, Jackman TR, Larson A. Niche lability in the evolution of a Caribbean lizard community. Nature. 2003;424:542–545. - PubMed
- Allen EE, Banfield JF. Community genomics in microbial ecology and evolution. Nat. Rev. Microbiol. 2005;3:489–498. - PubMed
- Kolter R, Greenberg EP. Microbial sciences: the superficial life of microbes. Nature. 2006;441:300–302. - PubMed
- Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol. Rev. 2009;33:206–224. - PubMed
- West SA, Diggle PD, Buckling A, Gardner A, Griffin AS. The Social lives of microbes. Annu. Rev. Ecol. Evol. Systemtatics. 2007;38:53–77.