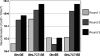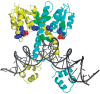Directed evolution of recombinase specificity by split gene reassembly - PubMed (original) (raw)
Directed evolution of recombinase specificity by split gene reassembly
Charles A Gersbach et al. Nucleic Acids Res. 2010 Jul.
Abstract
The engineering of new enzymes that efficiently and specifically modify DNA sequences is necessary for the development of enhanced gene therapies and genetic studies. To address this need, we developed a robust strategy for evolving site-specific recombinases with novel substrate specificities. In this system, recombinase variants are selected for activity on new substrates based on enzyme-mediated reassembly of the gene encoding beta-lactamase that confers ampicillin resistance to Escherichia coli. This stringent evolution method was used to alter the specificities of catalytic domains in the context of a modular zinc finger-recombinase fusion protein. Gene reassembly was detectable over several orders of magnitude, which allowed for tunable selectivity and exceptional sensitivity. Engineered recombinases were evolved to react with sequences from the human genome with only three rounds of selection. Many of the evolved residues, selected from a randomly-mutated library, were conserved among other members of this family of recombinases. This enhanced evolution system will translate recombinase engineering and genome editing into a practical and expedient endeavor for academic, industrial and clinical applications.
Figures
Figure 1.
Recombinase-dependent split gene reassembly system. (A) Structure of evolution vector before and after recombinase-mediated excision of the GFPuv fragment, with recombination target sites shown in blue. SpeI and HindIII restriction sites were inserted into the β-lactamase gene between Leu196 and Leu197 for convenient insertion of a GFPuv transgene flanked by recombination sites. The structure of a RecZF target site consists of a 20 bp core sequence flanked by zinc finger-binding sites (ZFBS). The C4 ZFBS was used in all plasmids and has been described previously (14). The sequences of various 20-bp core sequences used in this study are shown, including sequences derived from the natural Gin invertase and Tn3 resolvase target sites (20G and 20T, respectively), and a 20-bp sequence derived from the promoter of ErbB2 on human chromosome 17 (20E). (B) The predicted sequences of the 19-amino acid peptide that is grafted into the reassembled β-lactamase protein after excision of GFPuv are shown for the four evolution vectors used in this study. (C) Structure of TEM-1 β-lactamase. The N-terminal fragment is shown in orange, and the C-terminal fragment is shown in blue. The arrow indicates the insertion site of the peptide encoded by the recombination site, between Leu 196 and Leu 197 in the loop connecting helices 9 and 10.
Figure 2.
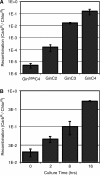
Gene reassembly of an evolution vector containing two C.20G sites (GG) by RecZFs containing a Gin catalytic domain was measured as the fraction of carbenicillin-resistant E. coli colonies (carbenicillin is an ampicillin analogue). (A) Recombination activity was dependent on the number of zinc finger domains on the RecZF, with four fingers showing more activity than two or three (P < 9_E_–7). Carbenicillin resistance in the presence of the catalytically inactive GinS9AC4 mutant represents the background level of recombination in this system. (B) Recombination activity by the RecZF with four fingers, GinC4, was also dependent on time in culture (P < 7_E_–5).
Figure 3.
(A) Cyclical selection strategy for the directed evolution of SSRs by split gene reassembly. (B) The selectivity and detection limit of the evolution system were determined by diluting GinC4-containing plasmid into catalytically inactive GinS9AC4 plasmid and performing two rounds of selection. The split gene reassembly system was capable of recovering one active GinC4 enzyme per 106 inactive GinS9AC4 enzymes.
Figure 4.
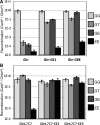
Directed evolution of recombinase specificity by split gene reassembly. Libraries of Gin and GinL7C7 variants underwent three rounds of selection with evolution vectors containing 20G and 20E (GE) or two 20E (EE) recombination sites. Activity of the library on the corresponding substrate increased with each round, except for Gin on the EE plasmid (Gin:EE).
Figure 5.
Recombination activity of the parent and evolved RecZF variants on diverse substrate sequences. The non-mutated starting clone and two unique variants from round 3 output libraries of (A) Gin selected on GE (variants GE1 & GE8) and (B) GinL7C7 selected on EE (variants EE2 & EE3) were tested for activity on vectors containing recombination sites of combinations of natural and unnatural core sequences. _P_-values with respect to vector & enzyme are (A) P < 4E–15, P < 2E-10 and (B) P < 6E–6, P < 3E–3.
Figure 6.
Residues evolved to conserved mutations from Gin:GE (blue; N11, N14, M70) and GinL7C7:EE (green; V6, F104, M108, I120) evolutions mapped onto the structure of a DNA-bound γδ resolvase dimer (yellow and cyan) (32). The catalytic serine (S9) is highlighted in red and DNA depicted in gray.
Figure 7.
Selected mutations are conserved among the family of serine recombinases. The name of the recombinase and associated accession number are shown on the left. The top row shows the wild-type Gin sequence and the second row shows selected mutations in this study (boxed) that share homology to other family members at that position (shaded).
Similar articles
- Expanding the zinc-finger recombinase repertoire: directed evolution and mutational analysis of serine recombinase specificity determinants.
Sirk SJ, Gaj T, Jonsson A, Mercer AC, Barbas CF 3rd. Sirk SJ, et al. Nucleic Acids Res. 2014 Apr;42(7):4755-66. doi: 10.1093/nar/gkt1389. Epub 2014 Jan 21. Nucleic Acids Res. 2014. PMID: 24452803 Free PMC article. - Evolution of programmable zinc finger-recombinases with activity in human cells.
Gordley RM, Smith JD, Gräslund T, Barbas CF 3rd. Gordley RM, et al. J Mol Biol. 2007 Mar 30;367(3):802-13. doi: 10.1016/j.jmb.2007.01.017. Epub 2007 Jan 12. J Mol Biol. 2007. PMID: 17289078 - Enhancing the specificity of recombinase-mediated genome engineering through dimer interface redesign.
Gaj T, Sirk SJ, Tingle RD, Mercer AC, Wallen MC, Barbas CF 3rd. Gaj T, et al. J Am Chem Soc. 2014 Apr 2;136(13):5047-56. doi: 10.1021/ja4130059. Epub 2014 Mar 20. J Am Chem Soc. 2014. PMID: 24611715 Free PMC article. - Serine recombinases as tools for genome engineering.
Brown WR, Lee NC, Xu Z, Smith MC. Brown WR, et al. Methods. 2011 Apr;53(4):372-9. doi: 10.1016/j.ymeth.2010.12.031. Epub 2010 Dec 30. Methods. 2011. PMID: 21195181 Review. - Site-specific DNA recombinases as instruments for genomic surgery.
Akopian A, Marshall Stark W. Akopian A, et al. Adv Genet. 2005;55:1-23. doi: 10.1016/S0065-2660(05)55001-6. Adv Genet. 2005. PMID: 16291210 Review.
Cited by
- Lipase improvement: goals and strategies.
Bassegoda A, Cesarini S, Diaz P. Bassegoda A, et al. Comput Struct Biotechnol J. 2012 Oct 15;2:e201209005. doi: 10.5936/csbj.201209005. eCollection 2012. Comput Struct Biotechnol J. 2012. PMID: 24688646 Free PMC article. Review. No abstract available. - Gene targeting to the ROSA26 locus directed by engineered zinc finger nucleases.
Perez-Pinera P, Ousterout DG, Brown MT, Gersbach CA. Perez-Pinera P, et al. Nucleic Acids Res. 2012 Apr;40(8):3741-52. doi: 10.1093/nar/gkr1214. Epub 2011 Dec 14. Nucleic Acids Res. 2012. PMID: 22169954 Free PMC article. - Targeted plasmid integration into the human genome by an engineered zinc-finger recombinase.
Gersbach CA, Gaj T, Gordley RM, Mercer AC, Barbas CF 3rd. Gersbach CA, et al. Nucleic Acids Res. 2011 Sep 1;39(17):7868-78. doi: 10.1093/nar/gkr421. Epub 2011 Jun 7. Nucleic Acids Res. 2011. PMID: 21653554 Free PMC article. - Structure-guided reprogramming of serine recombinase DNA sequence specificity.
Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF 3rd. Gaj T, et al. Proc Natl Acad Sci U S A. 2011 Jan 11;108(2):498-503. doi: 10.1073/pnas.1014214108. Epub 2010 Dec 27. Proc Natl Acad Sci U S A. 2011. PMID: 21187418 Free PMC article. - A programmable Cas9-serine recombinase fusion protein that operates on DNA sequences in mammalian cells.
Chaikind B, Bessen JL, Thompson DB, Hu JH, Liu DR. Chaikind B, et al. Nucleic Acids Res. 2016 Nov 16;44(20):9758-9770. doi: 10.1093/nar/gkw707. Epub 2016 Aug 11. Nucleic Acids Res. 2016. PMID: 27515511 Free PMC article.
References
- Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. - PubMed
- Akopian A, Marshall Stark W. Site-specific DNA recombinases as instruments for genomic surgery. Adv. Genet. 2005;55:1–23. - PubMed
- Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. - PubMed
- Liu Y, Lakshmipathy U, Ozgenc A, Thyagarajan B, Lieu P, Fontes A, Xue H, Scheyhing K, MacArthur C, Chesnut JD. hESC engineering by integrase-mediated chromosomal targeting. Methods Mol. Biol. 584:229–268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources