MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis - PubMed (original) (raw)
MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis
Lars Grøntved et al. Mol Cell Biol. 2010 May.
Abstract
The Mediator subunit MED1/TRAP220/DRIP205/PBP interacts directly with many nuclear receptors and was long thought to be responsible for tethering Mediator to peroxisome proliferator-activated receptor (PPAR)-responsive promoters. However, it was demonstrated recently that PPARgamma can recruit Mediator by MED1-independent mechanisms. Here, we show that target gene activation by ectopically expressed PPARgamma and PPARalpha is independent of MED1. Consistent with this finding, recruitment of PPARgamma, MED6, MED8, TATA box-binding protein (TBP), and RNA polymerase II (RNAPII) to the enhancer and proximal promoter of the PPARgamma target gene Fabp4 is also independent of MED1. Using a small interfering RNA (siRNA)-based approach, we identify MED14 as a novel critical Mediator component for PPARgamma-dependent transactivation, and we demonstrate that MED14 interacts directly with the N terminus of PPARgamma in a ligand-independent manner. Interestingly, MED14 knockdown does not affect the recruitment of PPARgamma, MED6, and MED8 to the Fabp4 enhancer but does reduce their occupancy of the Fabp4 proximal promoter. In agreement with the necessity of MED14 for PPARgamma transcriptional activity, we show that knockdown of MED14 impairs adipogenesis of 3T3-L1 cells. Thus, MED14 constitutes a novel anchoring point between Mediator and the N-terminal domain of PPARgamma that is necessary for functional PPARgamma-mediated recruitment of Mediator and transactivation of PPARgamma subtype-specific target genes.
Figures
FIG. 1.
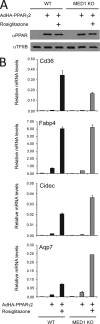
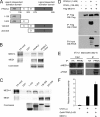
PPARγ activates target genes independently of MED1. WT and MED1 KO MEFs were transduced with AdHA-PPARγ2 in the presence or absence of 1 μM rosiglitazone. Whole-cell lysates and RNA were prepared 8 h after transduction. (A) Proteins from whole-cell extracts were separated by SDS-PAGE and analyzed by immunoblotting with antibodies against PPAR and TFIIB. α, anti. (B) RNA was quantified by real-time PCR with primers against Gtf2b, Fabp4, Cd36, Cidec, or Aqp7. Threshold cycle (CT) values were normalized to CT values from Gtf2b and visualized as relative mRNA levels. Error bars indicate the ranges of the results of experiments performed in duplicate. Results are representative of a minimum of three independent experiments.
FIG. 2.
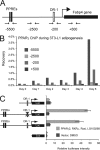
PPARγ occupies the proximal promoter of the Fabp4 gene indirectly. (A) Representation of the Fabp4 gene loci with the relative position of the reported PPREs. Primer pairs for ChIP analysis are indicated below the diagram. Numbers refer to positions relative to TSS. (B) ChIP-PCR confirmation of PPARγ occupancy of the proximal promoter of the Fabp4 gene. ChIP was performed during adipogenesis of 3T3-L1 adipocytes. PPAR occupancy of the Fabp4 enhancer and proximal promoter was investigated by real-time PCR with the indicated primers. (C) NIH 3T3 cells were transfected with the indicated luciferase (Luc) reporter constructs [pGL3basic-Fabp4(−4500/+1), pGL3basic-Fabp4(−7900/+1)DR1mut, pGL3basic-Fabp4(−7900/+1), and pGL3basic] together with pShuttle-CMV-PPARγ2, pShuttle-CMV-RXRα, and SV40 β-galactosidase in the presence or absence of 1 μM rosiglitazone (Rosi) and 200 nM LG100268. Luciferase and β-galactosidase activities were measured 24 h posttransfection, and luciferase activity was plotted as relative luciferase activity normalized to β-galactosidase activity. Data are represented as means ± standard errors of three independent experiments performed in triplicate. DMSO, dimethyl sulfoxide.
FIG. 3.
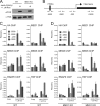
PPARγ-induced recruitment of Mediator at the Fabp4 promoter and enhancer is independent of MED1. WT and MED1 KO MEFs were transduced with adenovirus without an insert (empty) or AdHA-PPARγ2 in the presence of 1 μM rosiglitazone (Rosi). Whole-cell extracts and chromatin were prepared 8 h after transduction. (A) Proteins from whole-cell extracts were separated by SDS-PAGE and analyzed by immunoblotting with antibodies against PPAR and TFIIB. α, anti. (B) Representation of the Fabp4 gene loci and of the relative positions of the primer pairs for ChIP-PCR analysis are indicated below the diagram. (C and D) ChIP was performed using antibodies against the HA tag, TBP, RNAPII, MED1, MED6, and MED8. Enriched DNA was quantified using real-time PCR with the primer pairs (shown in panel B) indicated and plotted as the amount of DNA recovered relative to the amount of quantified DNA from input chromatin. Error bars indicate the ranges of the results of experiments performed in duplicates. Results are representative of a minimum of two independent experiments.
FIG. 4.
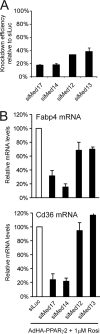
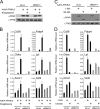
Knockdown of specific Mediator subunits reduces the transcriptional activity of PPARγ. WT MEFs were transfected with siRNA (20 nM) against luciferase (Luc), MED17, MED14, MED12, or MED13 for 24 h, followed by transduction with AdHA-PPARγ2 in the presence of 1 μM rosiglitazone (Rosi). (A) Knockdown efficiency of the different siRNAs against Mediator subunits relative to results for cells transfected with siRNA against luciferase. RNA expression levels were quantified by real-time PCR, normalized to the level of Gtf2b, and visualized as the percentage of the expression level in cells transfected with siRNA against luciferase. (B) RNA levels of the PPARγ target genes Cd36 and Fabp4 relative to that of Gtf2b were determined by real-time PCR and visualized as the percentage of the expression level in cells transfected with siRNA against luciferase. Error bars indicate the ranges of the results of experiments performed in duplicates. Results are representative of a minimum of three independent experiments.
FIG. 5.
MED14 interacts with the N-terminal domain of PPARγ. (A) PPARγ consists of four functional domains. The N-terminal A/B domain (amino acids [aa] 1 to 138) contains the ligand-independent transactivation function, the C (aa 138 to 203) and D (aa 203 to 279) domains are involved in DNA binding, and the E domain (aa 279 to 505) constitutes the ligand binding domain and has ligand-dependent transactivity. (B) MED14 interacts with GST-tagged full-length PPARγ2 in a ligand-independent manner. _In vitro_-translated and 35S-labeled MED14 and MED1 were incubated with immobilized GST-tagged PPARγ2 in the presence or absence of 1 μM rosiglitazone (Rosi). Bound proteins were separated by SDS-PAGE. (C) MED14 specifically interacts with the N-terminal domain of PPARγ2 _in vitro. In vitro_-translated and 35S-labeled MED14 and MED1 were incubated with immobilized GST-tagged PPARγ domains (represented in panel A) in the presence of 1 μM rosiglitazone. Bound proteins were separated by SDS-PAGE. Similar amounts of the GST-tagged PPARγ domains were verified by Coomassie staining. (D) MED14 interaction with PPARγ in cells is dependent on the N-terminal domain of PPARγ. Flag-tagged MED14, PPARγ2, or N-terminally truncated PPARγ was transfected into 239T cells, and cellular extracts were immunoprecipitated with antibody against the Flag epitope. Immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting with antibodies against Flag and PPAR. Amounts of 5% of the cellular extracts from transfected cells were immunoblotted with PPAR antibody to verify similar levels of expression of full-length and truncated PPARγ. WB, Western blot. (E) Endogenous PPARγ interacts with endogenous MED14 in 3T3-L1 adipocytes. Nuclear extracts from 3T3-L1 day 6 adipocytes were used for immunoprecipitation with antibody against HA or PPARγ. Immunoprecipitates were separated by SDS-PAGE (right) together with 10% input nuclear extract (left) and immunoblotted with antibodies against MED14 or PPARγ. α, anti. (F) MED14 coactivates the N-terminal transactivation domain of PPARγ. MEFs were transfected with a Gal4-responsive luciferase reporter (UAS-Luc) and a plasmid encoding the N-terminal A/B domain of PPARγ2 fused to the Gal4 DNA binding domain in the presence or absence of an expression plasmid encoding MED14. SV40 β-galactosidase was used for normalization. Error bars represent standard deviations of the results of experiments performed in triplicates. All results are representative of a minimum of two independent experiments.
FIG. 6.
Knockdown of MED14 compromises the ability of both full-length and A/B domain-truncated PPARγ to activate a subset of target genes. WT MEFs were transfected with siRNA (50 nM) against luciferase (Luc) or MED14 and, 24 h thereafter, transduced with AdHA-PPARγ2 (expressing full-length PPARγ2 [amino acids 1 to 505]) or AdHA-PPARγ-CDE (expressing truncated PPARγ [amino acids 138 to 505]), respectively, in the absence or presence of 1 μM rosiglitazone (Rosi). (A and C) Eight hours after adenoviral transduction, whole-cell extracts were prepared and analyzed by immunoblotting with antibodies against PPAR and TFIIB. α, anti. (B and D) RNA was prepared 8 h after transduction, and RNA levels of Gtf2b, Fabp4, Cidec, Cd36, Lpl, Acox1, and Cpt2 were determined by real-time PCR, normalized to the levels of Gtf2b, and visualized as relative mRNA levels. Error bars represent standard deviations of results of experiments performed in triplicates. Results are representative of a minimum of three independent experiments.
FIG. 7.
MED14 is necessary for Mediator recruitment to the proximal promoter of Fabp4. WT MEFs were transfected with siRNA (50 nM) against luciferase or MED14 and, 24 h thereafter, transduced with AdHA-PPARγ2 in the presence of 1 μM rosiglitazone. Whole-cell extracts and chromatin were prepared 8 h after adenoviral transduction. (A) Proteins from whole-cell extracts were separated by SDS-PAGE and analyzed by immunoblotting with antibodies against PPAR and TFIIB. α, anti. (B) ChIP-PCR was used to determine the relative levels of occupancy of PPARγ, MED1, RNAPII, MED6, and MED8 in the Fabp4 locus. Relative levels of occupancy, i.e., levels recovered with ChIP-PCR from cells overexpressing PPARγ2 relative to levels recovered from untransduced cells, are indicated. The figure shows the mean results of two independent experiments, with the ranges indicated by error bars. (C) In cells with normal expression of MED14, Mediator is recruited to an enhancer occupied by PPARγ:RXR through direct interaction between MED14 and the N-terminal A/B domain of PPARγ and between MED1 and the ligand binding domain (LBD) of PPARγ. This facilitates juxtaposition of the enhancer with the proximal promoter and, subsequently, recruitment of RNAPII (Pol II) to the promoter. In the absence of MED14, residual Mediator can still be recruited to the enhancer; however, functional interaction with the proximal promoter and recruitment of RNAPII is compromised.
FIG. 8.
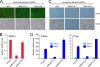
MED14 is essential for adipogenesis of 3T3-L1 cells. Proliferating 3T3-L1 cells were transduced with lentivirus expressing shRNAs targeting either LacZ or MED14. Cells were allowed to proliferate for 4 days and were then cultured to confluence and induced to differentiate. (A) Confluent undifferentiated cells were analyzed by epifluorescence microscopy to detect GFP expression. (B) RNA levels of MED14 in undifferentiated cells were determined by real-time PCR and normalized to that of Gtf2b. Error bars indicate the ranges of the results of experiments performed in duplicates. (C) Cells were stained with oil red O to visualize the accumulation of lipid droplets at days 0 and 6 of differentiation. (D) RNA levels of Gtf2b, Fabp4, and _Ppar_γ were determined by real-time PCR at day 0 and day 6 of differentiation. Levels were normalized to levels of Gtf2b and visualized as relative mRNA levels. Error bars indicate the ranges of the results of experiments performed in duplicates. Results are representative of a minimum of two independent experiments.
FIG. 9.
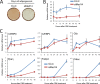
Preconfluent 3T3-L1 cells were transfected with siRNA against luciferase or MED14. Transfected cells were grown to confluence and subsequently differentiated into adipocytes by treatment with a cocktail of dexamethasone, methyl-isobutyl xanthine, and insulin (DMI). (A) Lipid accumulation was evaluated by oil red O staining at day 6 of differentiation. (B and C) RNA was purified 0, 6, 12, 24, and 48 h after addition of the adipogenic cocktail, and RNA expression levels of Gtf2b and Med14 (B) or C/EBPβ, _C/EBP_δ, Gilz, _PPAR_γ, Fabp4, and Cidec (C) were determined by real-time PCR. Levels were normalized to levels of Gtf2b and visualized as relative mRNA levels. Error bars indicate the ranges of the results of experiments performed in duplicates. Results are representative of two independent experiments.
Similar articles
- Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression.
Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. Ge K, et al. Mol Cell Biol. 2008 Feb;28(3):1081-91. doi: 10.1128/MCB.00967-07. Epub 2007 Nov 26. Mol Cell Biol. 2008. PMID: 18039840 Free PMC article. - The PPARgamma2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment.
Bugge A, Grøntved L, Aagaard MM, Borup R, Mandrup S. Bugge A, et al. Mol Endocrinol. 2009 Jun;23(6):794-808. doi: 10.1210/me.2008-0236. Epub 2009 Mar 12. Mol Endocrinol. 2009. PMID: 19282365 Free PMC article. - MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor.
Chen W, Rogatsky I, Garabedian MJ. Chen W, et al. Mol Endocrinol. 2006 Mar;20(3):560-72. doi: 10.1210/me.2005-0318. Epub 2005 Oct 20. Mol Endocrinol. 2006. PMID: 16239257 - PPARγ and the global map of adipogenesis and beyond.
Lefterova MI, Haakonsson AK, Lazar MA, Mandrup S. Lefterova MI, et al. Trends Endocrinol Metab. 2014 Jun;25(6):293-302. doi: 10.1016/j.tem.2014.04.001. Epub 2014 Apr 29. Trends Endocrinol Metab. 2014. PMID: 24793638 Free PMC article. Review. - Modulation of the transcriptional activity of peroxisome proliferator-activated receptor gamma by protein-protein interactions and post-translational modifications.
Kim TH, Kim MY, Jo SH, Park JM, Ahn YH. Kim TH, et al. Yonsei Med J. 2013 May 1;54(3):545-59. doi: 10.3349/ymj.2013.54.3.545. Yonsei Med J. 2013. PMID: 23549795 Free PMC article. Review.
Cited by
- Critical roles of transcriptional coactivator MED1 in the formation and function of mouse adipose tissues.
Ito K, Schneeberger M, Gerber A, Jishage M, Marchildon F, Maganti AV, Cohen P, Friedman JM, Roeder RG. Ito K, et al. Genes Dev. 2021 May 1;35(9-10):729-748. doi: 10.1101/gad.346791.120. Epub 2021 Apr 22. Genes Dev. 2021. PMID: 33888560 Free PMC article. - The transcription factor NKX1-2 promotes adipogenesis and may contribute to a balance between adipocyte and osteoblast differentiation.
Chen N, Schill RL, O'Donnell M, Xu K, Bagchi DP, MacDougald OA, Koenig RJ, Xu B. Chen N, et al. J Biol Chem. 2019 Nov 29;294(48):18408-18420. doi: 10.1074/jbc.RA119.007967. Epub 2019 Oct 15. J Biol Chem. 2019. PMID: 31615896 Free PMC article. - Unique and overlapping roles of NRF2 and NRF1 in transcriptional regulation.
Sekine H, Motohashi H. Sekine H, et al. J Clin Biochem Nutr. 2024 Mar;74(2):91-96. doi: 10.3164/jcbn.23-106. Epub 2023 Nov 22. J Clin Biochem Nutr. 2024. PMID: 38510688 Free PMC article. - Cyclin C regulates adipogenesis by stimulating transcriptional activity of CCAAT/enhancer-binding protein α.
Song Z, Xiaoli AM, Zhang Q, Zhang Y, Yang EST, Wang S, Chang R, Zhang ZD, Yang G, Strich R, Pessin JE, Yang F. Song Z, et al. J Biol Chem. 2017 May 26;292(21):8918-8932. doi: 10.1074/jbc.M117.776229. Epub 2017 Mar 28. J Biol Chem. 2017. PMID: 28351837 Free PMC article. - Involvement of CREB-regulated transcription coactivators (CRTC) in transcriptional activation of steroidogenic acute regulatory protein (Star) by ACTH.
Smith LIF, Huang V, Olah M, Trinh L, Liu Y, Hazell G, Conway-Campbell B, Zhao Z, Martinez A, Lefrançois-Martinez AM, Lightman S, Spiga F, Aguilera G. Smith LIF, et al. Mol Cell Endocrinol. 2020 Jan 1;499:110612. doi: 10.1016/j.mce.2019.110612. Epub 2019 Oct 8. Mol Cell Endocrinol. 2020. PMID: 31604124 Free PMC article.
References
- Aoyama, T., J. M. Peters, N. Iritani, T. Nakajima, K. Furihata, T. Hashimoto, and F. J. Gonzalez. 1998. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J. Biol. Chem. 273:5678-5684. - PubMed
- Barbera, M. J., A. Schluter, N. Pedraza, R. Iglesias, F. Villarroya, and M. Giralt. 2001. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene: a link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J. Biol. Chem. 276:1486-1493. - PubMed
- Chawla, A., W. A. Boisvert, C. H. Lee, B. A. Laffitte, Y. Barak, S. B. Joseph, D. Liao, L. Nagy, P. A. Edwards, L. K. Curtiss, R. M. Evans, and P. Tontonoz. 2001. A PPAR[gamma]-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 7:161-171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous