Complete mitochondrial genome of a Pleistocene jawbone unveils the origin of polar bear - PubMed (original) (raw)
. 2010 Mar 16;107(11):5053-7.
doi: 10.1073/pnas.0914266107. Epub 2010 Mar 1.
Stephan C Schuster, Yazhou Sun, Sandra L Talbot, Ji Qi, Aakrosh Ratan, Lynn P Tomsho, Lindsay Kasson, Eve Zeyl, Jon Aars, Webb Miller, Olafur Ingólfsson, Lutz Bachmann, Oystein Wiig
Affiliations
- PMID: 20194737
- PMCID: PMC2841953
- DOI: 10.1073/pnas.0914266107
Complete mitochondrial genome of a Pleistocene jawbone unveils the origin of polar bear
Charlotte Lindqvist et al. Proc Natl Acad Sci U S A. 2010.
Erratum in
- Proc Natl Acad Sci U S A. 2010 Mar 30;107(13):6118
Abstract
The polar bear has become the flagship species in the climate-change discussion. However, little is known about how past climate impacted its evolution and persistence, given an extremely poor fossil record. Although it is undisputed from analyses of mitochondrial (mt) DNA that polar bears constitute a lineage within the genetic diversity of brown bears, timing estimates of their divergence have differed considerably. Using next-generation sequencing technology, we have generated a complete, high-quality mt genome from a stratigraphically validated 130,000- to 110,000-year-old polar bear jawbone. In addition, six mt genomes were generated of extant polar bears from Alaska and brown bears from the Admiralty and Baranof islands of the Alexander Archipelago of southeastern Alaska and Kodiak Island. We show that the phylogenetic position of the ancient polar bear lies almost directly at the branching point between polar bears and brown bears, elucidating a unique morphologically and molecularly documented fossil link between living mammal species. Molecular dating and stable isotope analyses also show that by very early in their evolutionary history, polar bears were already inhabitants of the Artic sea ice and had adapted very rapidly to their current and unique ecology at the top of the Arctic marine food chain. As such, polar bears provide an excellent example of evolutionary opportunism within a widespread mammalian lineage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
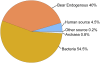
Fig. 1.
Metagenomic composition of the 454 sequence reads from the Poolepynten polar bear tooth. The pie chart shows the percentages of sequence reads assigned to the categories bacteria, bear endogenous, human, Archaea, and other sources. A comparison of the 454 sequence reads against the dog genome suggested that ∼40% of the reads represent endogenous bear DNA, whereas 4.5% and the remaining ∼55% represent human and bacterial contamination, respectively.
Fig. 2.
Mitogenomic sequence variation and organization. Sequence differences found among 17 bear mitochondrial genomes with respect to a previously published polar bear genome (GenBank accession no. NC_003428). Each vertical bar depicts a nucleotide difference from this reference sequence (shown at the bottom illustrating the organization of the genome into the different regions). The ancient Poolepynten bear sequence is highlighted in red.
Fig. 3.
Phylogenetic and chronographic reconstruction of polar bear evolution. (A) Maximum clade probability tree inferred from a BEAST analysis of complete mt genome sequences excluding the VNTR repeat in the D loop (
Table S2
). Numbers at selected nodes indicate mean ages in million years. The red bars at nodes illustrate the age width of the 95% highest posterior density interval. The posterior probability values of each clade are indicated in orange. An identical tree topology was obtained using maximum parsimony and maximum likelihood (bootstrap support values in green and yellow, respectively). For details on voucher information for the mtDNA genome sequences included in this study, see
Table S1
and
Figs. S3
and
S4
. The 2009 Geologic Time Scale with major relevant epochs is shown above the tree. (B) Phylogenetic network of complete mt genomes (excluding the VNTR repeat) of 11 polar and brown bears based on Neighbor-Net analysis with LogDet distances (see scale bar).
Similar articles
- Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears.
Cahill JA, Stirling I, Kistler L, Salamzade R, Ersmark E, Fulton TL, Stiller M, Green RE, Shapiro B. Cahill JA, et al. Mol Ecol. 2015 Mar;24(6):1205-17. doi: 10.1111/mec.13038. Epub 2015 Feb 5. Mol Ecol. 2015. PMID: 25490862 Free PMC article. - Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change.
Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE, Tomsho LP, Ibarra-Laclette E, Herrera-Estrella L, Peacock E, Farley S, Sage GK, Rode K, Obbard M, Montiel R, Bachmann L, Ingólfsson O, Aars J, Mailund T, Wiig O, Talbot SL, Lindqvist C. Miller W, et al. Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):E2382-90. doi: 10.1073/pnas.1210506109. Epub 2012 Jul 23. Proc Natl Acad Sci U S A. 2012. PMID: 22826254 Free PMC article. - Insights into bear evolution from a Pleistocene polar bear genome.
Lan T, Leppälä K, Tomlin C, Talbot SL, Sage GK, Farley SD, Shideler RT, Bachmann L, Wiig Ø, Albert VA, Salojärvi J, Mailund T, Drautz-Moses DI, Schuster SC, Herrera-Estrella L, Lindqvist C. Lan T, et al. Proc Natl Acad Sci U S A. 2022 Jun 14;119(24):e2200016119. doi: 10.1073/pnas.2200016119. Epub 2022 Jun 6. Proc Natl Acad Sci U S A. 2022. PMID: 35666863 Free PMC article. - Polar bears: the fate of an icon.
Fitzgerald KT. Fitzgerald KT. Top Companion Anim Med. 2013 Nov;28(4):135-42. doi: 10.1053/j.tcam.2013.09.007. Top Companion Anim Med. 2013. PMID: 24331553 Review. - Physiological consequences of Arctic sea ice loss on large marine carnivores: unique responses by polar bears and narwhals.
Pagano AM, Williams TM. Pagano AM, et al. J Exp Biol. 2021 Feb 24;224(Pt Suppl 1):jeb228049. doi: 10.1242/jeb.228049. J Exp Biol. 2021. PMID: 33627459 Review.
Cited by
- Ancestral polymorphisms and sex-biased migration shaped the demographic history of brown bears and polar bears.
Nakagome S, Mano S, Hasegawa M. Nakagome S, et al. PLoS One. 2013 Nov 13;8(11):e78813. doi: 10.1371/journal.pone.0078813. eCollection 2013. PLoS One. 2013. PMID: 24236053 Free PMC article. - Biomechanical consequences of rapid evolution in the polar bear lineage.
Slater GJ, Figueirido B, Louis L, Yang P, Van Valkenburgh B. Slater GJ, et al. PLoS One. 2010 Nov 5;5(11):e13870. doi: 10.1371/journal.pone.0013870. PLoS One. 2010. PMID: 21079768 Free PMC article. - Pros and cons of methylation-based enrichment methods for ancient DNA.
Seguin-Orlando A, Gamba C, Sarkissian C, Ermini L, Louvel G, Boulygina E, Sokolov A, Nedoluzhko A, Lorenzen ED, Lopez P, McDonald HG, Scott E, Tikhonov A, Stafford TW Jr, Alfarhan AH, Alquraishi SA, Al-Rasheid KAS, Shapiro B, Willerslev E, Prokhortchouk E, Orlando L. Seguin-Orlando A, et al. Sci Rep. 2015 Jul 2;5:11826. doi: 10.1038/srep11826. Sci Rep. 2015. PMID: 26134828 Free PMC article. - Evolutionary history of enigmatic bears in the Tibetan Plateau-Himalaya region and the identity of the yeti.
Lan T, Gill S, Bellemain E, Bischof R, Nawaz MA, Lindqvist C. Lan T, et al. Proc Biol Sci. 2017 Dec 13;284(1868):20171804. doi: 10.1098/rspb.2017.1804. Proc Biol Sci. 2017. PMID: 29187630 Free PMC article. - Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads--a baiting and iterative mapping approach.
Hahn C, Bachmann L, Chevreux B. Hahn C, et al. Nucleic Acids Res. 2013 Jul;41(13):e129. doi: 10.1093/nar/gkt371. Epub 2013 May 9. Nucleic Acids Res. 2013. PMID: 23661685 Free PMC article.
References
- Powell A, Shennan S, Thomas MG. Late Pleistocene demography and the appearance of modern human behavior. Science. 2009;324:1298–1301. - PubMed
- Kurtén B. The evolution of the polar bear, Ursus maritimus Phipps. Acta Zool Fenn. 1964;108:1–30.
- Shields GF, et al. Phylogeography of mitochondrial DNA variation in brown bears and polar bears. Mol Phylogenet Evol. 2000;15:319–326. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous