A primer on metagenomics - PubMed (original) (raw)
Review
A primer on metagenomics
John C Wooley et al. PLoS Comput Biol. 2010.
Abstract
Metagenomics is a discipline that enables the genomic study of uncultured microorganisms. Faster, cheaper sequencing technologies and the ability to sequence uncultured microbes sampled directly from their habitats are expanding and transforming our view of the microbial world. Distilling meaningful information from the millions of new genomic sequences presents a serious challenge to bioinformaticians. In cultured microbes, the genomic data come from a single clone, making sequence assembly and annotation tractable. In metagenomics, the data come from heterogeneous microbial communities, sometimes containing more than 10,000 species, with the sequence data being noisy and partial. From sampling, to assembly, to gene calling and function prediction, bioinformatics faces new demands in interpreting voluminous, noisy, and often partial sequence data. Although metagenomics is a relative newcomer to science, the past few years have seen an explosion in computational methods applied to metagenomic-based research. It is therefore not within the scope of this article to provide an exhaustive review. Rather, we provide here a concise yet comprehensive introduction to the current computational requirements presented by metagenomics, and review the recent progress made. We also note whether there is software that implements any of the methods presented here, and briefly review its utility. Nevertheless, it would be useful if readers of this article would avail themselves of the comment section provided by this journal, and relate their own experiences. Finally, the last section of this article provides a few representative studies illustrating different facets of recent scientific discoveries made using metagenomics.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Environmental Shotgun Sequencing (ESS).
(A) Sampling from habitat; (B) filtering particles, typically by size; (C) DNA extraction and lysis; (D) cloning and library; (E) sequence the clones; (F) sequence assembly.
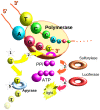
Figure 2. Pyrosequencing.
Single stranded DNA template is first hybridized with the sequencing primer and mixed with the enzymes along with the two substrates adenosine 5′-phosphosulfate (APS) and luciferin. In each cycle, (1) one of the four nucleotides (dTTPi, in this case) is then added to the reaction. (2) If the nucleotide is complementary to the base in the template strand then the DNA polymerase incorporates it into the growing strand. (3) Pyrophosphate (PPi)—in an amount equal in molarity to that of the incorporated nucleotide—is released and converted to ATP by sulfurylase in the presence of APS. (4) ATP then serves as a substrate to luciferase, causing a light reaction. Photon emission is in equimolar quanta to the amount of nucleotide incorporated in a given cycle. (5) The excess nucleotides are degraded by apyrase.
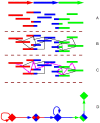
Figure 3. Fragment assembly.
(A–C) Hamiltonian. (A) A sequence with overlapping reads; (B) Each read is represented as a vertex, with edges connecting the overlapping vertices; (C) the assembly solution is a Hamiltonian path (all vertices are visited, no vertex is visited more than once) through the resulting graph; (D) For short reads assembly, each vertex is a _k_-mer (or a hashed collection of _k_-mers), and the reads are threaded between vertices as edges. The solution is a Eulerian path, where each edge is visited once. Repeats are merged into a single edge. For detailed algorithms see , , –.
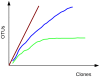
Figure 4. Rarefaction curves.
Green, most or all species have been sampled; blue, this habitat has not been exhaustively sampled; red, species rich habitat, only a small fraction has been sampled.
Similar articles
- An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review. - Applying Genome-Resolved Metagenomics to Deconvolute the Halophilic Microbiome.
Uritskiy G, DiRuggiero J. Uritskiy G, et al. Genes (Basel). 2019 Mar 14;10(3):220. doi: 10.3390/genes10030220. Genes (Basel). 2019. PMID: 30875864 Free PMC article. Review. - MG-RAST, a Metagenomics Service for Analysis of Microbial Community Structure and Function.
Keegan KP, Glass EM, Meyer F. Keegan KP, et al. Methods Mol Biol. 2016;1399:207-33. doi: 10.1007/978-1-4939-3369-3_13. Methods Mol Biol. 2016. PMID: 26791506 - Metagenomic analysis of uncultured microorganisms and their enzymatic attributes.
Bilal T, Malik B, Hakeem KR. Bilal T, et al. J Microbiol Methods. 2018 Dec;155:65-69. doi: 10.1016/j.mimet.2018.11.014. Epub 2018 Nov 16. J Microbiol Methods. 2018. PMID: 30452938 Review. - Single-cell metagenomics: challenges and applications.
Xu Y, Zhao F. Xu Y, et al. Protein Cell. 2018 May;9(5):501-510. doi: 10.1007/s13238-018-0544-5. Epub 2018 Apr 25. Protein Cell. 2018. PMID: 29696589 Free PMC article. Review.
Cited by
- Towards a predictive systems-level model of the human microbiome: progress, challenges, and opportunities.
Greenblum S, Chiu HC, Levy R, Carr R, Borenstein E. Greenblum S, et al. Curr Opin Biotechnol. 2013 Aug;24(4):810-20. doi: 10.1016/j.copbio.2013.04.001. Epub 2013 Apr 23. Curr Opin Biotechnol. 2013. PMID: 23623295 Free PMC article. Review. - Systematic genomic analysis reveals the complementary aerobic and anaerobic respiration capacities of the human gut microbiota.
Ravcheev DA, Thiele I. Ravcheev DA, et al. Front Microbiol. 2014 Dec 5;5:674. doi: 10.3389/fmicb.2014.00674. eCollection 2014. Front Microbiol. 2014. PMID: 25538694 Free PMC article. - Differences in sequencing technologies improve the retrieval of anammox bacterial genome from metagenomes.
Gori F, Tringe SG, Folino G, van Hijum SA, Op den Camp HJ, Jetten MS, Marchiori E. Gori F, et al. BMC Genomics. 2013 Jan 16;14:7. doi: 10.1186/1471-2164-14-7. BMC Genomics. 2013. PMID: 23324532 Free PMC article. - Genomic approaches to studying the human microbiota.
Weinstock GM. Weinstock GM. Nature. 2012 Sep 13;489(7415):250-6. doi: 10.1038/nature11553. Nature. 2012. PMID: 22972298 Free PMC article. Review. - Selection of models for the analysis of risk-factor trees: leveraging biological knowledge to mine large sets of risk factors with application to microbiome data.
Zhang Q, Abel H, Wells A, Lenzini P, Gomez F, Province MA, Templeton AA, Weinstock GM, Salzman NH, Borecki IB. Zhang Q, et al. Bioinformatics. 2015 May 15;31(10):1607-13. doi: 10.1093/bioinformatics/btu855. Epub 2015 Jan 6. Bioinformatics. 2015. PMID: 25568281 Free PMC article.
References
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. - PubMed
- Berg R. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. - PubMed
- Collins FS, McKusick VA. Implications of the human genome project for medical science. JAMA. 2001;285:540–544. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous