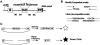Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells - PubMed (original) (raw)
Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells
Farjana Fattah et al. PLoS Genet. 2010.
Abstract
The repair of DNA double-strand breaks (DSBs) is critical for the maintenance of genomic integrity and viability for all organisms. Mammals have evolved at least two genetically discrete ways to mediate DNA DSB repair: homologous recombination (HR) and non-homologous end joining (NHEJ). In mammalian cells, most DSBs are preferentially repaired by NHEJ. Recent work has demonstrated that NHEJ consists of at least two sub-pathways-the main Ku heterodimer-dependent or "classic" NHEJ (C-NHEJ) pathway and an "alternative" NHEJ (A-NHEJ) pathway, which usually generates microhomology-mediated signatures at repair junctions. In our study, recombinant adeno-associated virus knockout vectors were utilized to construct a series of isogenic human somatic cell lines deficient in the core C-NHEJ factors (Ku, DNA-PK(cs), XLF, and LIGIV), and the resulting cell lines were characterized for their ability to carry out DNA DSB repair. The absence of DNA-PK(cs), XLF, or LIGIV resulted in cell lines that were profoundly impaired in DNA DSB repair activity. Unexpectedly, Ku86-null cells showed wild-type levels of DNA DSB repair activity that was dominated by microhomology joining events indicative of A-NHEJ. Importantly, A-NHEJ DNA DSB repair activity could also be efficiently de-repressed in LIGIV-null and DNA-PK(cs)-null cells by subsequently reducing the level of Ku70. These studies demonstrate that in human cells C-NHEJ is the major DNA DSB repair pathway and they show that Ku is the critical C-NHEJ factor that regulates DNA NHEJ DSB pathway choice.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Reporter substrate for analysis of NHEJ.
A cartoon of the reporter construct (pEGFP-Pem1-Ad2). (A) The construct is essentially a GFP cassette whose expression is driven by CMV promoter and terminated by the SV40 polyA sequence. “G” is separated from “FP” by a 2.4 kb intron containing an exon (Ad) from adenovirus that is flanked by HindIII and I-SceI restriction sites. Splice donor (SD) and splice acceptor (SA) sites are shown. (B) Restriction sites used to introduce DSBs. Digestion with HindIII generates compatible cohesive ends. Because I-SceI has a nonpalindromic 18-bp recognition site, cleavage of the two inverted I-SceI sites generates incompatible ends. (C) Due to the presence of the Ad-exon into the middle of the Pem1 intron, the Ad exon is efficiently spliced into the middle of the GFP ORF, inactivating the GFP activity and thus making the starting substrate GFP negative. Both sides of the Ad exon have HindIII/I-SceI restriction sites. Cleavage with either of these endonucleases removes the Ad exon and upon successful intracellular plasmid circularization GFP expression is restored and can be quantitated by flow cytometry.
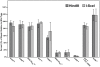
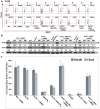
Figure 2. The impact of C-NHEJ mutations on end joining.
Four independent experiments comparable to those depicted in Figure S1 and Figure 4 were performed and the average percent repair relative to wild-type is shown with the standard deviation.
Figure 3. The loss of C-NHEJ greatly reduces end joining.
The indicated cell lines were transfected with _HindIII_- (Top panels) or I_-SceI_- (Bottom panels) linearized pEGFP-Pem1-Ad2 plasmid together with a pCherry plasmid. The cells were analyzed by FACS 24 hr post transfection. The number of cells that were doubly EGFP (horizontal) and pCherry (vertical) positive versus the number that were pCherry positive was determined. For a given experiment the data are shown as percent repair in the upper right-hand corner of each plot.
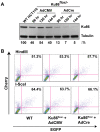
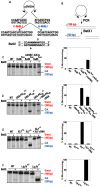
Figure 4. Ku86-null cells show wild-type levels of end joining activity.
(A) Western blot analysis shows that the expression of Cre (AdCre) in Ku86flox/− cells results in the reduction of Ku86 expression. AdCMV is a negative control adenoviral vector. (B) the indicated cell lines were transfected with either HindIII- (Top panels) or I-SceI- (bottom panels) linearized pEGFP-Pem1-Ad2 plasmid. All symbols are as in Figure 3.
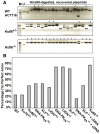
Figure 5. The absence of Ku86 results in predominately microhomology-based end joining.
(A) _HindIII_-linearized pEGFP-Pem1-Ad2 plasmids were recovered from either WT HCT116, Ku86+/− or Ku86−/− cells, propagated through E. coli and then analyzed for retention of a single HindIII restriction site (“perfect rejoining”) by HindIII restriction enzyme digestion analysis. The asterisks indicate those plasmids where perfect rejoining occurred. (B) The results of four or more experiments similar to those depicted in (A) were combined and summarized. N.B. The re-expression of a WT DNA-PKcs or XLF cDNA (+cDNA) in their respective null cell lines reduced the frequency of perfect rejoining back to WT levels, confirming the specificity of the effect.
Figure 6. The reduction of Ku results in elevated levels of end joining in C-NHEJ mutant cell lines.
(A) FACS profiles using the pEGFP-Pem1-Ad2 reporter substrate are shown for the indicated cell lines. The profiles for the Top and Bottom panels were generated using _HindIII_- and _I-SceI_-linearized plasmids, respectively. The percent of the substrate that was repaired is shown in the right-hand corner of each profile. (B) Western blot analyses demonstrate a reduction in Ku protein levels. Western blots for extracts derived from the indicated cell lines are shown using antibodies against either Ku86, Ku70 or (as a loading control) tubulin (Tub). Each of the blots was quantitated using a phosphoimager and the level of a particular Ku subunit relative to the amount expressed in the parental cell line is indicated below each blot. (C) Four independent experiments comparable to those depicted in (A) were performed and the average percent repair is shown with the standard deviation.
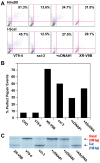
Figure 7. Independent confirmation of microhomology-mediated end joining in C-NHEJ mutant cell lines.
(A) Reporter substrate biased for use by microhomology-directed NHEJ (A-NHEJ). The reporter has been designed such that cleavage with Eco47III and EcoRV results in a blunt-ended linear substrate with 6-bp direct repeats (boxes) at both ends. C-NHEJ joining will result in the retention of some of both repeats whereas A-NHEJ should generate a single repeat, which is a substrate for BstXI. This figure is excerpted from Verkaik et al., 2002, Eur. J. Immunol., 32∶701. (B) The experimental scheme for analysis of the plasmids recovered from transfected cells. The plasmids were subjected to PCR using one radiolabeled (asterisk) primer. The PCR products were then subjected to BstXI restriction enzyme digestion. (C–F) Left Panels: Autoradiograms of representative microhomology assays using the indicated cell lines. The size of the primary PCR product (180 bp) and the BstXI cleavage product (120 bp) are indicated. Right Panels: Three independent experiments similar to the ones shown on the left were quantitated with a phosphoimager and averaged.
Figure 8. Microhomology-mediated end joining in hamster Ku86-null cell lines.
(A) The indicated cell lines were transfected with _HindIII_- (Top panels) or _I-SceI_-(Bottom panels) linearized pEGFP-Pem1-Ad2 together with a supercoiled pCherry plasmid (to monitor transfection efficiency). The number in the top right corner corresponds to the percentage of cells that turned green after 24 hr as a percentage of the cells productively transfected. +cDNA#1 corresponds to a sxi-3 cell line that has been stably complemented with a Ku86 cDNA. (B) _HindIII_-linearized pEGFP-Pem1-Ad2 plasmids were recovered from the indicated cell lines, propagated through E. coli and then analyzed for retention of a single HindIII restriction site (“perfect rejoining”) by HindIII restriction enzyme digestion analysis. (C) The indicated cell lines were transfected with the pDVG94 plasmid that had been linearized by Eco47III and EcoRV digestion. After 24 hr, the plasmids were recovered and then analyzed by PCR as described in Figure 7.
Similar articles
- Mechanisms of DNA double strand break repair and chromosome aberration formation.
Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G. Iliakis G, et al. Cytogenet Genome Res. 2004;104(1-4):14-20. doi: 10.1159/000077461. Cytogenet Genome Res. 2004. PMID: 15162010 Review. - The mutagenic potential of non-homologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV.
Kuhfittig-Kulle S, Feldmann E, Odersky A, Kuliczkowska A, Goedecke W, Eggert A, Pfeiffer P. Kuhfittig-Kulle S, et al. Mutagenesis. 2007 May;22(3):217-33. doi: 10.1093/mutage/gem007. Epub 2007 Mar 7. Mutagenesis. 2007. PMID: 17347130 - DNA ligase III and DNA ligase IV carry out genetically distinct forms of end joining in human somatic cells.
Oh S, Harvey A, Zimbric J, Wang Y, Nguyen T, Jackson PJ, Hendrickson EA. Oh S, et al. DNA Repair (Amst). 2014 Sep;21:97-110. doi: 10.1016/j.dnarep.2014.04.015. Epub 2014 May 16. DNA Repair (Amst). 2014. PMID: 24837021 Free PMC article. - Biochemical evidence for Ku-independent backup pathways of NHEJ.
Wang H, Perrault AR, Takeda Y, Qin W, Wang H, Iliakis G. Wang H, et al. Nucleic Acids Res. 2003 Sep 15;31(18):5377-88. doi: 10.1093/nar/gkg728. Nucleic Acids Res. 2003. PMID: 12954774 Free PMC article. - X-ray scattering reveals disordered linkers and dynamic interfaces in complexes and mechanisms for DNA double-strand break repair impacting cell and cancer biology.
Hammel M, Tainer JA. Hammel M, et al. Protein Sci. 2021 Sep;30(9):1735-1756. doi: 10.1002/pro.4133. Epub 2021 Jun 5. Protein Sci. 2021. PMID: 34056803 Free PMC article. Review.
Cited by
- KAP1 Deacetylation by SIRT1 Promotes Non-Homologous End-Joining Repair.
Lin YH, Yuan J, Pei H, Liu T, Ann DK, Lou Z. Lin YH, et al. PLoS One. 2015 Apr 23;10(4):e0123935. doi: 10.1371/journal.pone.0123935. eCollection 2015. PLoS One. 2015. PMID: 25905708 Free PMC article. - XRN2 Links Transcription Termination to DNA Damage and Replication Stress.
Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL, Boothman DA. Morales JC, et al. PLoS Genet. 2016 Jul 20;12(7):e1006107. doi: 10.1371/journal.pgen.1006107. eCollection 2016 Jul. PLoS Genet. 2016. PMID: 27437695 Free PMC article. - Targeting abnormal DNA repair in therapy-resistant breast cancers.
Tobin LA, Robert C, Nagaria P, Chumsri S, Twaddell W, Ioffe OB, Greco GE, Brodie AH, Tomkinson AE, Rassool FV. Tobin LA, et al. Mol Cancer Res. 2012 Jan;10(1):96-107. doi: 10.1158/1541-7786.MCR-11-0255. Epub 2011 Nov 23. Mol Cancer Res. 2012. PMID: 22112941 Free PMC article. - Current and Prospective Applications of CRISPR-Cas12a in Pluricellular Organisms.
Khan S, Sallard E. Khan S, et al. Mol Biotechnol. 2023 Feb;65(2):196-205. doi: 10.1007/s12033-022-00538-5. Epub 2022 Aug 8. Mol Biotechnol. 2023. PMID: 35939208 Free PMC article. Review. - Cancer risk at low doses of ionizing radiation: artificial neural networks inference from atomic bomb survivors.
Sasaki MS, Tachibana A, Takeda S. Sasaki MS, et al. J Radiat Res. 2014 May;55(3):391-406. doi: 10.1093/jrr/rrt133. Epub 2013 Dec 22. J Radiat Res. 2014. PMID: 24366315 Free PMC article. Review.
References
- Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–1976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA77598/CA/NCI NIH HHS/United States
- P30 CA077598/CA/NCI NIH HHS/United States
- R01 GM069576/GM/NIGMS NIH HHS/United States
- GM069576/GM/NIGMS NIH HHS/United States
- T32 AG029796/AG/NIA NIH HHS/United States
- HL079559/HL/NHLBI NIH HHS/United States
- R01 HL079559/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous