A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction - PubMed (original) (raw)
Review
A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction
Andrea F Moon et al. Protein Sci. 2010 May.
Abstract
Protein crystallographers are often confronted with recalcitrant proteins not readily crystallizable, or which crystallize in problematic forms. A variety of techniques have been used to surmount such obstacles: crystallization using carrier proteins or antibody complexes, chemical modification, surface entropy reduction, proteolytic digestion, and additive screening. Here we present a synergistic approach for successful crystallization of proteins that do not form diffraction quality crystals using conventional methods. This approach combines favorable aspects of carrier-driven crystallization with surface entropy reduction. We have generated a series of maltose binding protein (MBP) fusion constructs containing different surface mutations designed to reduce surface entropy and encourage crystal lattice formation. The MBP advantageously increases protein expression and solubility, and provides a streamlined purification protocol. Using this technique, we have successfully solved the structures of three unrelated proteins that were previously unattainable. This crystallization technique represents a valuable rescue strategy for protein structure solution when conventional methods fail.
Figures
Figure 1
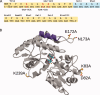
The tandem fixed-arm MBP/SER System. A: Schematic of the pMALX polylinker region. Sequences in yellow originated from the commercially available pMAL-c2X vector (New England Biolabs). Alanine substitutions in the C-terminal α-helix are highlighted in red with the nucleotide changes shown above or below in black. The sequence boxed in light blue shows the point of truncation of the MBP sequence after Asn367, deletion of the Factor Xa cleavage site, and insertion of NotI and NheI restriction sites. B: Ribbon diagram of the structure of MBP (PDB ID code
1HSJ
- showing the residues included in the crystallization cassettes. The C-terminal α-helix bearing three alanine substitutions is shown in purple. The bound maltose is drawn in stick (cyan). The positions of the residues that were substituted with alanine for the SER mutations are drawn in stick in orange. This figure was generated using MolScript.
Figure 2
Crystal-structure of MBP(wt)-2OST (PDB ID code
3F5F
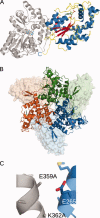
). The maltose bound to the MBP is drawn in stick in cyan. The PAP co-factor bound to the 2OST is drawn in stick in green. Ribbon diagrams were generated using PyMOL. A: Ribbon diagram of the crystal structure of the MBP(wt)-2OST fusion protein. The structure of the MBP is shown in gray. α-helices from the 2OST structure are shown in blue, β-strands are shown in red, and the random coil regions are shown in yellow. B: Ribbon diagram of the trimeric MBP(wt)-2OST. Molecule 1 is shown in green, Molecule 2 in blue, and Molecule 3 is shown in orange. 2OST molecules are drawn in ribbon and the MBP molecules are shown as surface renderings. C: Ribbon diagram of the MBP(wt)-2OST interaction surface. Secondary structural elements of the 2OST are shown in blue. The C-terminal α-helix of MBP is shown in gray, with the K359A and K362A mutations drawn in stick.
Figure 3
Crystal structure of the MBP(C)-RACK1A fusion protein (PDB ID code
3DM0
). A: Ribbon diagram of the MBP(C)-RACK1A fusion protein. The bound maltose is drawn in stick (cyan). The structure of the MBP is shown in gray, relative to that of the RACK1A (multicolored). The β-strands of the RACK1A β-propeller domain are shown as ribbons, with each WD motif colored and labeled. The residues of the disordered loop (Lys277-Lys294) in WD6 (purple) are labeled. B,C, and D: Secondary structural elements of the MBP(C) protein are drawn as gray ribbons, with the SER mutations drawn in stick. The positions of symmetry-related MBP molecules (gray) and RACK1A (green, blue, yellow) are drawn in stick. B: Position of the D82A/K83A SER mutations in crystal lattice formation. C: Position of the K239A mutation in crystal lattice formation. D: Role of Asn173 in crystal lattice formation.
Figure 4
Crystal structure of the MBP(E)-Der p 7 fusion protein (PDB ID code
3H4Z
). A: The ribbon diagram of Molecule A of MBP(E)-Der p 7. The structure of the MBP is shown in gray, with the bound maltose drawn in stick (cyan). α-helices of the Der p 7 structure are shown in green, β-strands in dark blue, and random coil regions in yellow. B: Intermolecular interactions comprising the asymmetric unit of the MBP(E)-Der p 7 crystals. Surface renderings of the MBP molecules (Molecule A in light green, Molecule B in light purple, Molecule C in light gold) illustrating the extensive MBP-MBP interactions within the asymmetric unit. Minimal interactions between the Der p 7 of Molecule B (dark purple) with the MBP of Molecule C (yellow). C: Position of the K239A mutation in the crystal lattice. The K239A mutations of Molecules A (green) and B (purple) lie along an intermolecular interaction surface.
Similar articles
- Crystal structures of MBP fusion proteins.
Waugh DS. Waugh DS. Protein Sci. 2016 Mar;25(3):559-71. doi: 10.1002/pro.2863. Epub 2016 Jan 9. Protein Sci. 2016. PMID: 26682969 Free PMC article. Review. - MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Gumpena R, Lountos GT, Waugh DS. Gumpena R, et al. Acta Crystallogr F Struct Biol Commun. 2018 Sep 1;74(Pt 9):549-557. doi: 10.1107/S2053230X18009901. Epub 2018 Aug 29. Acta Crystallogr F Struct Biol Commun. 2018. PMID: 30198887 Free PMC article. - Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana.
Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. Ullah H, et al. Protein Sci. 2008 Oct;17(10):1771-80. doi: 10.1110/ps.035121.108. Epub 2008 Aug 20. Protein Sci. 2008. PMID: 18715992 Free PMC article. - Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein.
Center RJ, Kobe B, Wilson KA, Teh T, Howlett GJ, Kemp BE, Poumbourios P. Center RJ, et al. Protein Sci. 1998 Jul;7(7):1612-9. doi: 10.1002/pro.5560070715. Protein Sci. 1998. PMID: 9684894 Free PMC article. - Crystal structures of fusion proteins with large-affinity tags.
Smyth DR, Mrozkiewicz MK, McGrath WJ, Listwan P, Kobe B. Smyth DR, et al. Protein Sci. 2003 Jul;12(7):1313-22. doi: 10.1110/ps.0243403. Protein Sci. 2003. PMID: 12824478 Free PMC article. Review.
Cited by
- Structure-function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome.
Patrick AN, Cabrera JH, Smith AL, Chen XS, Ford HL, Zhao R. Patrick AN, et al. Nat Struct Mol Biol. 2013 Apr;20(4):447-53. doi: 10.1038/nsmb.2505. Epub 2013 Feb 24. Nat Struct Mol Biol. 2013. PMID: 23435380 Free PMC article. - Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity.
Mueller GA, Gosavi RA, Pomés A, Wünschmann S, Moon AF, London RE, Pedersen LC. Mueller GA, et al. Allergy. 2011 Jul;66(7):878-85. doi: 10.1111/j.1398-9995.2010.02532.x. Epub 2011 Jan 21. Allergy. 2011. PMID: 21255036 Free PMC article. - Crystal structure of the human dual specificity phosphatase 1 catalytic domain.
Gumpena R, Lountos GT, Raran-Kurussi S, Tropea JE, Cherry S, Waugh DS. Gumpena R, et al. Protein Sci. 2018 Feb;27(2):561-567. doi: 10.1002/pro.3328. Epub 2017 Nov 21. Protein Sci. 2018. PMID: 29052270 Free PMC article. - Highlighting the potential utility of MBP crystallization chaperone for Arabidopsis BIL1/BZR1 transcription factor-DNA complex.
Nosaki S, Terada T, Nakamura A, Hirabayashi K, Xu Y, Bui TBC, Nakano T, Tanokura M, Miyakawa T. Nosaki S, et al. Sci Rep. 2021 Feb 16;11(1):3879. doi: 10.1038/s41598-021-83532-2. Sci Rep. 2021. PMID: 33594119 Free PMC article. - A carrier protein strategy yields the structure of dalbavancin.
Economou NJ, Nahoum V, Weeks SD, Grasty KC, Zentner IJ, Townsend TM, Bhuiya MW, Cocklin S, Loll PJ. Economou NJ, et al. J Am Chem Soc. 2012 Mar 14;134(10):4637-45. doi: 10.1021/ja208755j. Epub 2012 Mar 1. J Am Chem Soc. 2012. PMID: 22352468 Free PMC article.
References
- Means GE, Feeney RE. Reductive alkylation of proteins. Anal Biochem. 1995;224:1–16. - PubMed
- Rayment I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 1997;276:171–179. - PubMed
- Hegy GB, Shackleton CH, Carlquist M, Bonn T, Engstrom O, Sjoholm P, Witkowska HE. Carboxymethylation of the human estrogen receptor ligand-binding domain-estradiol complex: HPLC/ESMS peptide mapping shows that cysteine 447 does not react with iodoacetic acid. Steroids. 1996;61:367–373. - PubMed
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous