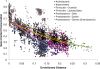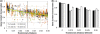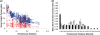Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives - PubMed (original) (raw)
Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives
Jesse R Zaneveld et al. Nucleic Acids Res. 2010 Jul.
Abstract
The mammalian gut is an attractive model for exploring the general question of how habitat impacts the evolution of gene content. Therefore, we have characterized the relationship between 16 S rRNA gene sequence similarity and overall levels of gene conservation in four groups of species: gut specialists and cosmopolitans, each of which can be divided into pathogens and non-pathogens. At short phylogenetic distances, specialist or cosmopolitan bacteria found in the gut share fewer genes than is typical for genomes that come from non-gut environments, but at longer phylogenetic distances gut bacteria are more similar to each other than are genomes at equivalent evolutionary distances from non-gut environments, suggesting a pattern of short-term specialization but long-term convergence. Moreover, this pattern is observed in both pathogens and non-pathogens, and can even be seen in the plasmids carried by gut bacteria. This observation is consistent with the finding that, despite considerable interpersonal variation in species content, there is surprising functional convergence in the microbiome of different humans. Finally, we observe that even within bacterial species or genera 16S rRNA divergence provides useful information about average conservation of gene content. The results described here should be useful for guiding strain selection to maximize novel gene discovery in large-scale genome sequencing projects, while the approach could be applied in studies seeking to understand the effects of habitat adaptation on genome evolution across other body habitats or environment types.
Figures
Figure 1.
Classification of species by habitat and pathogenicity. (a) All genomes for the Actinobacteria, Bacteroidetes, Firmicutes (separating the Clostridiales and the Lactobacillales), δ-Proteobacteria, ε-Proteobacteria, and the γ-Proteobacteria (Enterobacteria) present in the KEGG database were downloaded (195 genomes total). The genomes were classified as follows (see ‘Materials and Methods’ section for detailed description): (i) BLAST was used to compare 16 S rRNA sequences for each genome against the NCBI Envs database to determine the environmental distribution of the species. (ii) Genomes were characterized by examination of the study titles of hits: genomes found exclusively in gut or fecal samples were labeled ‘gut specialist’, those found in several studies of the gut, but also in other environments were categorized as ‘gut cosmopolitan’, while those never found in the gut were labeled ‘non-gut’. (iii) In borderline cases where genomes were found in several environmental samples and only a small number of gut samples, isolation information from the GOLD database was used to determine whether the genome should be categorized as ‘gut cosmopolitan’ or ‘non-gut’. Probiotic bacteria, or those isolated from the gastrointestinal tract or feces in this abundance class were taken to be ‘gut cosmopolitan’. (iv) Finally, genomes in each category were categorized by pathogenicity using the GOLD (26) annotations for ‘phenotype’ and ‘disease’. Commensal microbes capable of only opportunistic infection were treated as non-pathogens in this analysis. Additionally, 13 genomes where annotation information was ambiguous or conflicted with observations from 16 S rRNA observations were removed from the analysis. (b) Example output of this annotation process, and numbers of genomes in each subcategory. Abbreviations are as follows: ‘G’, gut specialist, ‘GC’ cosmopolitan resident of the gut, ‘N’ non-gut. Pathogens are denoted ‘P’ and non-pathogens ‘N’.
Figure 2.
Gene conservation by evolutionary distance. Gene content conservation at the protein level. Each point represents a BLAST comparison between two genomes at an _E_-value threshold cutoff of 10−10. The _x_-axis represents the 16 S distance between the two genomes, while the _y_-axis represents the proportion of proteins from the query genome that matches proteins from the subject genome. Genome–genome comparisons are subdivided by taxonomic group. Comparisons between members of the same taxonomic group are represented by the same shape and similar colors. Each colored line represents the exponential regression of the points within a single taxon. _r_2 values for exponential regression of each taxon were: Actinobacteria, _r_2 = 0.28; Bacteroidetes, _r_2 = 0.70; Clostridia, _r_2 = 0.57; Lactobacillales, _r_2 = 0.70; δ-Proteobacteria, _r_2 = 0.38; ε-Proteobacteria _r_2 = 0.48; γ-Proteobacteria _r_2 = 0.24.
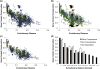
Figure 3.
Gene conservation in gut-adapted bacteria. Relationship between evolutionary distance in terms of 16 S rRNA divergence and gene content conservation. For these graphs, the _x_-axis shows evolutionary divergences in terms of nucleotide substitutions per site in the 16S rRNA gene, and the _y_-axis shows the fraction of genes in the first species that are found in the second species using BLASTP on the translated sequences. (a) Each point represents a comparison between two genomes. Yellow points are comparisons between two genomes that are both gut specialists, green points are comparisons between two genomes that are both cosmopolitan members of the gut microbiota, wheras all other comparisons are considered together and colored in blue. Although much variation in gene conservation is explained by phylogenetic distance, examples of genomes that vary little or greatly in gene conservation can be found at any given distance. _r_2 = 0.82 for gut specialists; 0.80 for gut cosmopolitans; and 0.22 for other comparisons. (b) Effects of relative genome size on conservation of gene content (size categories are defined in ‘Materials and Methods’ section above). Genome–genome comparisons were plotted separately for pairs of genomes where both are in the same size category (blue squares), where one genome is medium and the other is either large or small (green squares), or where one genome is large and the other is small (yellow squares). (c) Gene content conservation in pairs of gut-adapted bacteria with similar genome sizes. When only gut specialist or gut cosmpolitan genomes are considered, and when both genomes in each pair are similarly sized, phylogenetic distance is predictive of gene content conservation: _r_2 = 0.81 gut specialists; 0.78 gut cosmopolitan; and 0.57 for other comparisons. (d) Depicts the same data as in (c), but binned into increments of 0.03 corrected substitutions per site in the 16S rRNA, to clarify trends in conservation. Specialist (white bars) and cosmopolitan (gray bars) bacteria inhabiting the gut have somewhat lower levels of gene conservation at evolutionary distances below 0.03 substitutions per site than non-gut bacteria (black bars), but elevated levels between ∼0.06–0.18 substitutions per site. Error bars depict standard error.
Figure 4.
Greater 16 S rRNA divergence implies greater divergence in gene content within bacterial species. (a) Trees constructed from either the full length 16 S rRNA or 250 nucleotide stretches of its V2, V4 or V6 regions. The vertical bar corresponds to the species boundary, using the traditional bacterial species definition of >97% 16 S rRNA identity. (This boundary was determined by regressing the corrected 16 S rRNA distances displayed here against 16 S rRNA percent identity. See
Supplementary Figure S2
). The results demonstrate that even within the same bacterial species, the average gene conservation of a genome pair falls as phylogenetic distance increases. (b) Binning the results from (a) to bins of 0.015 16 S rRNA substitutions per site allows quantification of the effects of phylogenetic distance on gene conservation. Black bars represent average gene conservation at a given distance when distances are calculated using the full-length 16 S rRNA gene sequence, while progressively lighter gray bars represent gene conservation when calculating distance with fragments of the V2, V4 or V6 regions, respectively.
Figure 5.
Gene conservation in plasmids borne by gut-adapted bacteria. (a) Gene conservation in bacterial chromosomes (red squares) or plasmids (blue squares). Plasmids show both lower average gene conservation than bacterial chromosomes, and, as would be expected given frequent conjugative exchange, a weaker relationship between evolutionary distance and gene conservation (_r_2 = 0.60 genomes; _r_2 = 0.06 plasmids). (b) Plasmids borne by specialist (white bars) or cosmopolitan (gray bars) bacteria tend to have higher gene conservation at evolutionary distances between 0.09 and 0.21 16 S rRNA substitutions per site than those borne by non-gut bacteria (black bars). These plasmids also exhibit markedly reduced gene conservation at distances under 0.03 substitutions per site.
Figure 6.
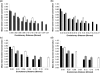
Gut pathogens, like gut commensals, exhibit different patterns of gene content conservation from non-gut genomes. Each panel depicts average levels of gene content conservation, binned in ranges of 0.03 16 S rRNA substitutions per site. Values for comparisons between pairs of non-gut bacteria are shown in black, pairs of gut cosmopolitan bacteria in gray and pairs of gut specialists in white. (a) Gene conservation in non-pathogens, including comparison between pairs in all size categories. (b) As in (a), but showing only comparisons between pairs of genomes in the same size category. (c) As in (a), but for pathogenic bacteria. (d) As in (b), but for pathogens. Error bars depict the standard error of the mean.
References
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008759/GM/NIGMS NIH HHS/United States
- R01HG004872/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P01DK078669/DK/NIDDK NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
- T32 GM08759/GM/NIGMS NIH HHS/United States