New primers for promising single-copy genes in fungal phylogenetics and systematics - PubMed (original) (raw)
doi: 10.3767/003158509X470602. Epub 2009 Aug 4.
A Crespo, P K Divakar, J D Fankhauser, E Herman-Sackett, K Kalb, M P Nelsen, N A Nelson, E Rivas-Plata, A D Shimp, T Widhelm, H T Lumbsch
Affiliations
- PMID: 20198159
- PMCID: PMC2802727
- DOI: 10.3767/003158509X470602
New primers for promising single-copy genes in fungal phylogenetics and systematics
I Schmitt et al. Persoonia. 2009 Dec.
Abstract
Developing powerful phylogenetic markers is a key concern in fungal phylogenetics. Here we report degenerate primers that amplify the single-copy genes Mcm7 (MS456) and Tsr1 (MS277) across a wide range of Pezizomycotina (Ascomycota). Phylogenetic analyses of 59 taxa belonging to the Eurotiomycetes, Lecanoromycetes, Leotiomycetes, Lichinomycetes and Sordariomycetes, indicate the utility of these loci for fungal phylogenetics at taxonomic levels ranging from genus to class. We also tested the new primers in silico using sequences of Saccharomycotina, Taphrinomycotina and Basidiomycota to predict their potential of amplifying widely across the Fungi. The analyses suggest that the new primers will need no, or only minor sequence modifications to amplify Saccharomycotina, Taphrinomycotina and Basidiomycota.
Keywords: Ascomycota; DNA replication licensing factor; MS277; MS456; Mcm7; Tsr1; evolution; lichenised fungi; phylogeny; pre-rRNA processing protein; protein-coding.
Figures
Fig. 1
Locations of the new primers for Mcm7 and Tsr1 using Aspergillus nidulans mRNA (XM_658504 and XM_658778) as reference sequence. Shaded areas in Tsr1 indicate regions of high sequence variability.
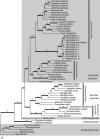
Fig. 2
Phylogeny of Pezizomycotina (Ascomycota) based on a combined alignment of Mcm7 (MS456) and Tsr1 (MS277) sequences. Total alignment length is 1203 bp. This is a 50 % majority rule consensus tree based on a sampling of 108 000 B/MCMC trees. Bold branches indicate posterior probabilities ≥ 0.95. Numbers above branches are maximum parsimony bootstrap support values ≥ 70 based on 2 000 random addition replicates.
Fig. 3
Comparison of the new primers to homologous sequences in Saccharomycotina (Ashbya, Kluyveromyces, Saccharomyces, Yarrowia), Taphrinomycotina (Schizosaccharomyces) and Basidiomycota (Coprinopsis, Cryptococcus, Ustilago). 100 % matches between primer sequence and gene sequences studied are indicated by grey shading. High sequence similarities indicate that the new primers are likely to fit in members of the analysed groups. Some primer sequences may require slight modifications.
Similar articles
- Examining new phylogenetic markers to uncover the evolutionary history of early-diverging fungi: comparing MCM7, TSR1 and rRNA genes for single- and multi-gene analyses of the Kickxellomycotina.
Tretter ED, Johnson EM, Wang Y, Kandel P, White MM. Tretter ED, et al. Persoonia. 2013 Jun;30:106-25. doi: 10.3767/003158513X666394. Epub 2013 Mar 20. Persoonia. 2013. PMID: 24027350 Free PMC article. - New molecular markers for fungal phylogenetics: two genes for species-level systematics in the Sordariomycetes (Ascomycota).
Walker DM, Castlebury LA, Rossman AY, White JF Jr. Walker DM, et al. Mol Phylogenet Evol. 2012 Sep;64(3):500-12. doi: 10.1016/j.ympev.2012.05.005. Epub 2012 May 22. Mol Phylogenet Evol. 2012. PMID: 22626621 - A multigene phylogenetic synthesis for the class Lecanoromycetes (Ascomycota): 1307 fungi representing 1139 infrageneric taxa, 317 genera and 66 families.
Miadlikowska J, Kauff F, Högnabba F, Oliver JC, Molnár K, Fraker E, Gaya E, Hafellner J, Hofstetter V, Gueidan C, Otálora MA, Hodkinson B, Kukwa M, Lücking R, Björk C, Sipman HJ, Burgaz AR, Thell A, Passo A, Myllys L, Goward T, Fernández-Brime S, Hestmark G, Lendemer J, Lumbsch HT, Schmull M, Schoch CL, Sérusiaux E, Maddison DR, Arnold AE, Lutzoni F, Stenroos S. Miadlikowska J, et al. Mol Phylogenet Evol. 2014 Oct;79:132-68. doi: 10.1016/j.ympev.2014.04.003. Epub 2014 Apr 18. Mol Phylogenet Evol. 2014. PMID: 24747130 Free PMC article. - Evolution of Mating in the Saccharomycotina.
Wolfe KH, Butler G. Wolfe KH, et al. Annu Rev Microbiol. 2017 Sep 8;71:197-214. doi: 10.1146/annurev-micro-090816-093403. Epub 2017 Jun 28. Annu Rev Microbiol. 2017. PMID: 28657889 Review. - The Fungal Tree of Life: from Molecular Systematics to Genome-Scale Phylogenies.
Spatafora JW, Aime MC, Grigoriev IV, Martin F, Stajich JE, Blackwell M. Spatafora JW, et al. Microbiol Spectr. 2017 Sep;5(5). doi: 10.1128/microbiolspec.FUNK-0053-2016. Microbiol Spectr. 2017. PMID: 28917057 Review.
Cited by
- New insights into the DNA extraction and PCR amplification of minute ascomycetes in the genus Laboulbenia (Pezizomycotina, Laboulbeniales).
Van Caenegem W, Haelewaters D. Van Caenegem W, et al. IMA Fungus. 2024 Jun 11;15(1):14. doi: 10.1186/s43008-024-00146-9. IMA Fungus. 2024. PMID: 38863065 Free PMC article. - A fifty-locus phylogenetic analysis provides deep insights into the phylogeny of Tricholoma (Tricholomataceae, Agaricales).
Ding XX, Xu X, Cui YY, Kost G, Wang PM, Yang ZL. Ding XX, et al. Persoonia. 2023 Jun;50:1-26. doi: 10.3767/persoonia.2023.50.01. Epub 2023 Jan 24. Persoonia. 2023. PMID: 38567264 Free PMC article. - Plant growth promotion under phosphate deficiency and improved phosphate acquisition by new fungal strain, Penicillium olsonii TLL1.
Suraby EJ, Agisha VN, Dhandapani S, Sng YH, Lim SH, Naqvi NI, Sarojam R, Yin Z, Park BS. Suraby EJ, et al. Front Microbiol. 2023 Oct 19;14:1285574. doi: 10.3389/fmicb.2023.1285574. eCollection 2023. Front Microbiol. 2023. PMID: 37965551 Free PMC article. - Phylogeny and species delimitations in the economically, medically, and ecologically important genus Samsoniella (Cordycipitaceae, Hypocreales).
Wang Y, Wang ZQ, Thanarut C, Dao VM, Wang YB, Yu H. Wang Y, et al. MycoKeys. 2023 Oct 3;99:227-250. doi: 10.3897/mycokeys.99.106474. eCollection 2023. MycoKeys. 2023. PMID: 37828936 Free PMC article. - Hesperomyces (Fungi, Ascomycota) associated with Hyperaspis ladybirds (Coleoptera, Coccinellidae): Rethinking host specificity.
Van Caenegem W, Ceryngier P, Romanowski J, Pfister DH, Haelewaters D. Van Caenegem W, et al. Front Fungal Biol. 2023 Jan 9;3:1040102. doi: 10.3389/ffunb.2022.1040102. eCollection 2022. Front Fungal Biol. 2023. PMID: 37746211 Free PMC article.
References
- Aguileta G, Marthey S, Chiapello H, Lebrun MH, Rodolphe F, Fournier E, Gendrault-Jacquemard A, Giraud T. 2008. Assessing the performance of single-copy genes for recovering robust phylogenies. Systematic Biology 57: 613 – 627 - PubMed
- Binder M, Hibbett DS. 2002. Higher-level phylogenetic relationships of homobasidiomycetes (mushroom-forming fungi) inferred from four rDNA regions. Molecular Phylogenetics and Evolution 22: 76 – 90 - PubMed
- Crespo A, Lumbsch HT, Mattsson JE, Blanco O, Divakar PK, Articus K, Wiklund E, Bawingan PA, Wedin M. 2007. Testing morphology-based hypotheses of phylogenetic relationships in Parmeliaceae (Ascomycota) using three ribosomal markers and the nuclear RPB1 gene. Molecular Phylogenetics and Evolution 44: 812 – 824 - PubMed
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783 – 791 - PubMed
- Gargas A, Depriest PT, Grube M, Tehler A. 1995. Multiple origins of lichen symbioses in Fungi suggested by SSU rDNA phylogeny. Science 268: 1492 – 1495 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources