Noninvasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells - PubMed (original) (raw)
. 2010 May;298(5):G582-9.
doi: 10.1152/ajpgi.00004.2010. Epub 2010 Mar 4.
Chen Zhao, Ivan Ivanov, Laurie A Davidson, Jennifer S Goldsby, Joanne R Lupton, Rose Ann Mathai, Marcia H Monaco, Deshanie Rai, W Michael Russell, Sharon M Donovan, Edward R Dougherty
Affiliations
- PMID: 20203060
- PMCID: PMC2867429
- DOI: 10.1152/ajpgi.00004.2010
Noninvasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells
Robert S Chapkin et al. Am J Physiol Gastrointest Liver Physiol. 2010 May.
Abstract
We have developed a novel molecular methodology that utilizes stool samples containing intact sloughed epithelial cells to quantify intestinal gene expression profiles in the developing human neonate. Since nutrition exerts a major role in regulating neonatal intestinal development and function, our goal was to identify gene sets (combinations) that are differentially regulated in response to infant feeding. For this purpose, fecal mRNA was isolated from exclusively breast-fed (n = 12) and formula-fed (n = 10) infants at 3 mo of age. Linear discriminant analysis was successfully used to identify the single genes and the two- to three-gene combinations that best distinguish the feeding groups. In addition, putative "master" regulatory genes were identified using coefficient of determination analysis. These results support our premise that mRNA isolated from stool has value in terms of characterizing the epigenetic mechanisms underlying the developmentally regulated transcriptional activation/repression of genes known to modulate gastrointestinal function. As larger data sets become available, this methodology can be extended to validation and, ultimately, identification of the main nutritional components that modulate intestinal maturation and function.
Figures
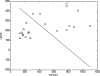
Fig. 1.
Linear discriminant analysis (LDA) phenotype classification using the genes endothelial PAS domain-containing protein 1 (hypoxia-inducible factor 2α, EPAS1) and uncoupling protein 2 (UCP2) provides the best-performing 2-gene feature set. Classification is between breast-fed (○) and formula-fed (△) infants (see Table 2 for additional details). Axes represent normalized intensity values of the indicated genes. Note clear separation between the groups, except for 1 outlier.
Fig. 2.
LDA phenotype classification using the genes EPAS1, forkhead box protein E3 (FOXE3), and synaptophysin (SYP) provides the best-performing 3-gene feature set. The 3-dimensional LDA hyperplane discriminates between breast-fed (○) and formula-fed (▵) infants (see Table 2 for additional details). Axes represent normalized intensity values of the corresponding genes. Note clear separation between the groups, except for 1 outlier (the same infant outlier in Fig. 1).
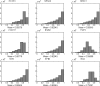
Fig. 3.
Association between expression patterns of genes (triple predictors) was determined using coefficient of determination (CoD). CoD measures the degree to which the transcriptional levels of an observed gene set can be used to improve the prediction of the transcriptional state of a target gene relative to the best-possible prediction in the absence of observations. Examples for the strong single-gene classifiers in Table 2, excluding tight junction protein 1 (TJP1), are shown. NR5A2, nuclear receptor subfamily 5, group A, member 2; NR3C1, nuclear receptor subfamily 3, group C, member 1; PCDH7, protocadherin 7; ITGB2, integrin-β2; EPIM, epimorphin; BAD, Bcl2 antagonist of cell death.
Similar articles
- Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides.
Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Donovan SM, et al. Adv Nutr. 2012 May 1;3(3):450S-5S. doi: 10.3945/an.112.001859. Adv Nutr. 2012. PMID: 22585924 Free PMC article. Review. - Recovery of exfoliated cells from the gastrointestinal tract of premature infants: a new tool to perform "noninvasive biopsies?".
Kaeffer B, des Robert C, Alexandre-Gouabau MC, Pagniez A, Legrand A, Amarger V, Küster A, Piloquet H, Champ M, le Huërou-Luron I, Rozé JC. Kaeffer B, et al. Pediatr Res. 2007 Nov;62(5):564-9. doi: 10.1203/PDR.0b013e318155a402. Pediatr Res. 2007. PMID: 17805197 - Fecal short-chain fatty acids of very-low-birth-weight preterm infants fed expressed breast milk or formula.
Pourcyrous M, Nolan VG, Goodwin A, Davis SL, Buddington RK. Pourcyrous M, et al. J Pediatr Gastroenterol Nutr. 2014 Dec;59(6):725-31. doi: 10.1097/MPG.0000000000000515. J Pediatr Gastroenterol Nutr. 2014. PMID: 25079478 - Human Breast Milk and Infant Formulas Differentially Modify the Intestinal Microbiota in Human Infants and Host Physiology in Rats.
Liu Z, Roy NC, Guo Y, Jia H, Ryan L, Samuelsson L, Thomas A, Plowman J, Clerens S, Day L, Young W. Liu Z, et al. J Nutr. 2016 Feb;146(2):191-9. doi: 10.3945/jn.115.223552. Epub 2015 Dec 16. J Nutr. 2016. PMID: 26674765 Clinical Trial. - Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects.
Le Huërou-Luron I, Blat S, Boudry G. Le Huërou-Luron I, et al. Nutr Res Rev. 2010 Jun;23(1):23-36. doi: 10.1017/S0954422410000065. Epub 2010 May 10. Nutr Res Rev. 2010. PMID: 20450531 Review.
Cited by
- Summary of the Joint National Institutes of Health and the Food and Drug Administration Workshop Titled "Exploring the Science Surrounding the Safe Use of Bioactive Ingredients in Infant Formula: Considerations for an Assessment Framework".
Donovan SM, Abrams SA, Azad MB, Belfort MB, Bode L, Carlson SE, Dallas DC, Hettinga K, Järvinen K, Kim JH, Lebrilla CB, McGuire MK, Sela DA, Neu J. Donovan SM, et al. J Pediatr. 2023 Apr;255:30-41.e1. doi: 10.1016/j.jpeds.2022.11.027. Epub 2022 Dec 2. J Pediatr. 2023. PMID: 36463938 Free PMC article. No abstract available. - Sepsis in preterm infants causes alterations in mucosal gene expression and microbiota profiles compared to non-septic twins.
Cernada M, Bäuerl C, Serna E, Collado MC, Martínez GP, Vento M. Cernada M, et al. Sci Rep. 2016 May 16;6:25497. doi: 10.1038/srep25497. Sci Rep. 2016. PMID: 27180802 Free PMC article. - Non-invasive Assessment of Fecal Stress Biomarkers in Hunting Dogs During Exercise and at Rest.
Zannoni A, Pietra M, Gaspardo A, Accorsi PA, Barone M, Turroni S, Laghi L, Zhu C, Brigidi P, Forni M. Zannoni A, et al. Front Vet Sci. 2020 Apr 21;7:126. doi: 10.3389/fvets.2020.00126. eCollection 2020. Front Vet Sci. 2020. PMID: 32373631 Free PMC article. - Colonic mucosal and exfoliome transcriptomic profiling and fecal microbiome response to a flaxseed lignan extract intervention in humans.
Lampe JW, Kim E, Levy L, Davidson LA, Goldsby JS, Miles FL, Navarro SL, Randolph TW, Zhao N, Ivanov I, Kaz AM, Damman C, Hockenbery DM, Hullar MAJ, Chapkin RS. Lampe JW, et al. Am J Clin Nutr. 2019 Aug 1;110(2):377-390. doi: 10.1093/ajcn/nqy325. Am J Clin Nutr. 2019. PMID: 31175806 Free PMC article. Clinical Trial. - Mapping gastrointestinal gene expression patterns in wild primates and humans via fecal RNA-seq.
Sharma AK, Pafčo B, Vlčková K, Červená B, Kreisinger J, Davison S, Beeri K, Fuh T, Leigh SR, Burns MB, Blekhman R, Petrželková KJ, Gomez A. Sharma AK, et al. BMC Genomics. 2019 Jun 14;20(1):493. doi: 10.1186/s12864-019-5813-z. BMC Genomics. 2019. PMID: 31200636 Free PMC article.
References
- Albaugh GP, Iyengar V, Lohani A, Malayeri M, Bala S, Nair P. Isolation of exfoliated colonic epithelial cells, a novel, non-invasive approach to the study of cellular markers. Int J Cancer 52: 347–350, 1992 - PubMed
- Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program 62: 13–29, 2008 - PubMed
- Braga-Neto UM, Dougherty ER. Bolstered error estimation. Pattern Recognition 37: 1267–1281, 2004
- Brenna JT, Salem N, Jr, Sinclair AJ, Cunnane SC. α-Linolenic acid supplementation and conversion to n-3 long-chain polyunsaturated fatty acids in humans. Prostaglandins Leukot Essent Fatty Acids 80: 85–91, 2009 - PubMed
- Burrin DB, Shulman RG, Reeds PJ, Davis TA, Gravitt KR. Porcine colostrum and milk stimulate visceral organ and skeletal muscle protein synthesis in neonatal piglets. J Nutr 122: 1205–1213, 1992 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA059034/CA/NCI NIH HHS/United States
- CA-59034/CA/NCI NIH HHS/United States
- CA-129444/CA/NCI NIH HHS/United States
- DK-71707/DK/NIDDK NIH HHS/United States
- P30 ES009106/ES/NIEHS NIH HHS/United States
- P30 ES-09106/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials