A nuclear factor-binding domain in the 5'-untranslated region of the amyloid precursor protein promoter: implications for the regulation of gene expression - PubMed (original) (raw)
A nuclear factor-binding domain in the 5'-untranslated region of the amyloid precursor protein promoter: implications for the regulation of gene expression
Alexander A Vostrov et al. BMC Res Notes. 2010.
Abstract
Background: The extracellular deposition of aggregated amyloid beta-protein is a neuropathological manifestation of Alzheimer disease and Down syndrome. The Amyloid beta-protein is derived from a group of larger differentially spliced proteins, the amyloid protein precursors (APP). Data suggests that the level of APP gene expression could contribute to the pathological processes leading to amyloid depositions.
Findings: The 5' untranslated region (UTR) of the APP gene, encompassing 147 base pairs between the transcriptional (+1) and the translational start site, was examined for its role in APP expression. Deletions close to the transcriptional start site reduced expression from the APP promoter in part by transcriptional mechanisms. However, deletions between position +50 and +104 had no effect on transcriptional activity while significantly reducing overall expression from the promoter. A nuclear factor-binding domain designated as DAPB was identified between position +72 and +115 of the 5'-APP-UTR. The binding-recognition sequence was localized between position +96 and +105. The same mutations that eliminated factor-binding also reduced expression from the APP promoter while having no effect on in vitro transcription or the RNA levels transcribed from transfected constructs.
Conclusions: A nuclear factor-binding domain designated as DAPB was identified in the 5'-UTR of the APP gene. Elimination of factor-binding correlated with an overall decline in expression from the APP promoter while in vitro transcription and the total amount of in vivo transcribed RNA remained unaffected. This suggests that the binding-factor may have a function in post-transcriptional regulation, including nuclear export of mRNA.
Figures
Figure 1
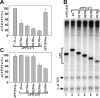
Structure of the APP promoter and the 5' UTR. (A) Schematic representation of the APP promoter with boxes indicating locations of regulatory regions and the DAPB domain discussed in the text. (B) The sequence of the 5'-_APP_-UTR from position +1 to +147. The translational start site (ATG) would follow position +147 in the native APP gene. Also shown are deletions at position +10, +30, +50, and +70 (arrows) and pertinent nucleotide positions as discussed in the text. Brackets show relevant restriction sites, of which SalI is part of the plasmid polylinker region. The sequence defining DNase I protected domain DAPB (solid lines) and the sequence recognized by the binding factor (dotted lines) are indicated by boxes. (C) Schematic representation of internal deletions D [10]-D [70] within the APP [147] fragment. Deleted sequences are indicated by dotted lines.
Figure 2
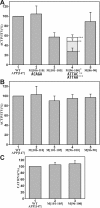
Expression from the APP promoter with deletions within the 5'-_APP_-UTR. (A) Expression from the full-length APP promoter and internal 5'-_APP_-UTR deletions by transient transfection in HeLa cells. The results are presented as CAT activities relative to the wild type APP [147] construct, which was assigned the value of 100% (column 1). Columns 2-5 show internal deletions D [70]-D [10] within APP [147] and column 6 shows the APP [104] construct that terminates at position +104. The results represent the average values of at least ten separate transfections with standard deviations (error bars). (B) Run-off in vitro transcription from promoter construct APP [104] (lane 1), APP [147] (lane 2) and sequential deletions D [70]-D [10] (lanes 3-6). The bracket delineates fragments transcribed from the APP promoter. Arrows indicate the two transcripts originating from the β-actin promoter. (C) Quantitation of transcription reactions illustrated in B. The APP transcripts were normalized to identical β-actin transcripts and the full-length (FL) APP [147] construct was assigned the value of 100% (column 2). Results represent the average of four independent experiments with standard deviations (error bars).
Figure 3
Characterization of the DAPB binding domain in the 5'-_APP_-UTR. (A) DNase I footprinting of a wild type APP promoter fragment extending from position -40 including the UTR to position +147 (lanes 1 and 6). The coding strand was 5' end-labeled with [32P] and the resulting fragment was digested with DNase I either in the presence (+) or absence (-) of nuclear extract. Brackets delineate the position of DNase I protected domain DAPB from position +72 to +115 as discussed in the text. (B) Mobility shift electrophoresis with elution fractions from SP ion exchange chromatography of whole HeLa cell nuclear extract and a [32P] 5' end-labeled double stranded oligonucleotide containing the 5'-_APP_-UTR sequence from position +60 to +120 [60-120]. The binding complex eluted predominantly in fractions 11-13 (lanes 3-5). The binding complex (b) and the free oligonucleotide (f) are indicated by brackets throughout the figure. (C) Mobility shift competition with the 5'-_APP_-UTR fragment [60-120] as a labeled probe: Mobility shift without competitor (lane 1); mobility shift with a 5-fold (lane 2) and 20-fold (lane 3) molar excess of unlabeled wild type [60-120] sequence; competition with 5-fold (lane 4), 20-fold (lane 5), and 100-fold (lane 6) excess of unlabeled oligonucleotide containing transverse mutations from position +86 to +100 (M [86-100]) within the [60-120] fragment (lower panel); competition with a 5-fold (lane 7), 20-fold (lane 8), and 100-fold (lane 9) excess of unlabeled oligonucleotide containing transverse mutations from position +71 to +85 (M [81-85]). (D) Mobility shift competition with the [60-120] fragment as a labeled probe (lane 1). Self-competition with a 10-fold (lane 2) and 40-fold (lane 3) excess of wild type [60-120] sequence and competition with a 40-fold unlabeled excess of mutations M [86-90] (lane 4), M [91-95] (lane 5), M [96-100] (lane 6), M [101-105] (lane 7), and M [86-100] (lane 8).
Figure 4
Expression from the APP promoter with mutations within the 5'-_APP_-UTR. (A) Transient transfection in HeLa cells. The activities of consecutive 5-base-pair block mutations M [106-110]-M [86-90](columns 2-5) are presented as CAT activities relative to the wild type APP [147] construct (column 1), which was assigned the value of 100%. In addition to the original M [96-100] mutation (column 4, *) [see Fig. 3D for sequence], the activities of two additional mutations (column 4, ** and ***) that removed the ATG sequence were investigated. The results represent the average values of at least ten separate transfections with standard deviations (error bars). (B) Run-off in vitro transcription in HeLa cell nuclear extract with the same designations as in A. (C) CAT RNA levels in HeLa cells transfected with constructs APP [147] WT (column 1), M [101-105] (column 2), and M [96-100] (column 3) were determined by quantitative RT-PCR. The wild type APP [147] construct (column 1) was assigned the value of 100% and the results represent the average of four independent experiments with standard deviations (error bars).
Similar articles
- Regulation and expression of the Alzheimer's beta/A4 amyloid protein precursor in health, disease, and Down's syndrome.
Beyreuther K, Pollwein P, Multhaup G, Mönning U, König G, Dyrks T, Schubert W, Masters CL. Beyreuther K, et al. Ann N Y Acad Sci. 1993 Sep 24;695:91-102. doi: 10.1111/j.1749-6632.1993.tb23035.x. Ann N Y Acad Sci. 1993. PMID: 8239320 Review. - Taking down the unindicted co-conspirators of amyloid beta-peptide-mediated neuronal death: shared gene regulation of BACE1 and APP genes interacting with CREB, Fe65 and YY1 transcription factors.
Lahiri DK, Ge YW, Rogers JT, Sambamurti K, Greig NH, Maloney B. Lahiri DK, et al. Curr Alzheimer Res. 2006 Dec;3(5):475-83. doi: 10.2174/156720506779025224. Curr Alzheimer Res. 2006. PMID: 17168646 Review.
Cited by
- The 5' untranslated region of potato SBgLR gene contributes to pollen-specific expression.
Chang Y, Yan M, Yu J, Zhu D, Zhao Q. Chang Y, et al. Planta. 2017 Sep;246(3):389-403. doi: 10.1007/s00425-017-2695-7. Epub 2017 Apr 25. Planta. 2017. PMID: 28444448 - Transcriptional and Post-Transcriptional Regulations of Amyloid-β Precursor Protein (APP) mRNA.
Sato K, Takayama KI, Hashimoto M, Inoue S. Sato K, et al. Front Aging. 2021 Aug 11;2:721579. doi: 10.3389/fragi.2021.721579. eCollection 2021. Front Aging. 2021. PMID: 35822056 Free PMC article. Review. - Advances in microRNA experimental approaches to study physiological regulation of gene products implicated in CNS disorders.
Long JM, Lahiri DK. Long JM, et al. Exp Neurol. 2012 Jun;235(2):402-18. doi: 10.1016/j.expneurol.2011.12.043. Epub 2012 Jan 5. Exp Neurol. 2012. PMID: 22245616 Free PMC article. Review. - HNF4α regulates claudin-7 protein expression during intestinal epithelial differentiation.
Farkas AE, Hilgarth RS, Capaldo CT, Gerner-Smidt C, Powell DR, Vertino PM, Koval M, Parkos CA, Nusrat A. Farkas AE, et al. Am J Pathol. 2015 Aug;185(8):2206-18. doi: 10.1016/j.ajpath.2015.04.023. Am J Pathol. 2015. PMID: 26216285 Free PMC article.
References
LinkOut - more resources
Full Text Sources