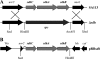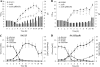Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms - PubMed (original) (raw)
Advantage of upregulation of succinate dehydrogenase in Staphylococcus aureus biofilms
Rosmarie Gaupp et al. J Bacteriol. 2010 May.
Abstract
Previous studies have demonstrated that various tricarboxylic acid (TCA) cycle genes, particularly the succinate dehydrogenase genes (sdhCAB), are upregulated in Staphylococcus aureus biofilms. To better study the role of this enzyme complex, an sdhCAB deletion mutant (Deltasdh) was constructed. Compared to the wild type (wt) the mutant was impaired in planktonic growth under aerobic conditions, excreted acetic acid could not be reused and accumulated continuously, succinate was excreted and found in the culture supernatant, and metabolome analysis with cells grown in chemically defined medium revealed reduced uptake/metabolism of some amino acids from the growth medium. Moreover, the mutant was able to counteract the steadily decreasing extracellular pH by increased urease activity. The addition of fumarate to the growth medium restored the wt phenotype. The mutant showed a small-colony variant (SCV)-like phenotype, a slight increase in resistance to various aminoglycoside antibiotics, and decreased pigmentation. The decreased growth under aerobic conditions is due to the interruption of the TCA cycle (indicated by the accumulation of succinate and acetic acid) with the consequence that many fewer reduction equivalents (NADH and FADH2) can fuel the respiratory chain. The results indicate that the TCA cycle is required for acetate and amino acid catabolism; its upregulation under biofilm conditions is advantageous under such nutrient- and oxygen-limited conditions.
Figures
FIG. 1.
Construction of the succinate dehydrogenase deletion mutant and complementation. (A) Location of the sdh operon in the chromosome of SA113 (upper panel) and the sdhCAB deletion mutant (Δ_sdh_; lower panel). (B) Gene arrangement in the complementation plasmid pRB_sdh_, including the putative native promoter region upstream (arrow). Plasmid-encoded genes: bla, beta-lactamase gene; ble, bleomycin resistance gene; cat, chloramphenicol resistance gene; uvrC, 3′ end of the sdh upstream gene.
FIG. 2.
Comparison of growth parameters and extracellular metabolites in S. aureus and the Δ_sdh_ mutant. (A and B) Growth curves and pH profile (bars) of SA113 (A) and HG001 (B). (C and D) Metabolite concentrations in the culture supernatant of SA113 (C) and its Δ_sdh_ mutant (D). Strains were aerobically cultivated in TSB at 37°C. Error bars indicate standard deviations from the means of three experiments.
FIG. 3.
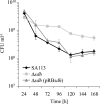
Long-term survival assay results. Strains were aerobically cultivated in TSB at 37°C for 7 days. Cultural aliquots were removed daily, spotted on TSB agar plates, and incubated at 37°C for 24 h. The values (CFU per milliliter) were determined by the drop plate method (17) in six replicates. Error bars indicate standard deviations from the means of five experiments.
FIG. 4.
Size of colonies on Mueller-Hinton agar plates. The SA113Δ_sdh_ mutant displayed a SCV phenotype. Supplementation of the agar with 5 mM glucose or 5 mM sodium fumarate, but not with 20 mM sodium succinate, restored wild-type colony size.
FIG. 5.
Absorption spectrum of extracted pigments. SA113 (A) and HG001 (B) along with corresponding Δ_sdh_ mutants were aerobically cultivated for 24 h in TSB. Absorption spectra were determined in ethanol extracts (2.4 μl ethanol per mg [wet weight] of cells). Error bars indicate standard deviations from the means of four experiments.
FIG. 6.
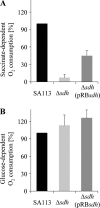
Polarographic measurement of the oxygen consumption rate, determined with a Clark-type oxygen electrode. SA113 and isogenic Δ_sdh_ mutants were grown aerobically for 20 to 24 h. Respiration of washed cell suspensions was initiated either with 100 mM succinate (A) or 1 mM glucose (B) as a substrate. The oxygen consumption rate of SA113 was set as 100%. Error bars indicate standard deviations from the means of three experiments.
FIG. 7.
Schematic representation of observed metabolic differences in the Δ_sdh_ mutant. In the Δ_sdh_ mutant urease activity was increased, succinate and acetate were excreted in higher amounts, the pH of the culture supernatant stayed low, and consumption of the amino acids alanine (ala), glutamate (glu), histidine (his), isoleucine (ile), and proline (pro) was affected. The corresponding genes of the indicated enzymatic reactions are annotated in S. aureus NCTC8325: ArcC, carbamate kinase (SAOUHSC_01129 and SAOUHSC_02965); ArgF, ornithine carbamoyltransferase (SAOUHSC_01128); ArgG, argininosuccinate synthase (SAOUHSC_00899); ArgH, argininiosuccinate lyase (SAOUHSC_00898); Arg, arginase (SAOUHSC_02409); UreA, urease subunit gamma (SAOUHSC_02558); UreB, urease subunit beta (SAOUHSC_02559); UreC, urease subunit alpha (SAOUHSC_02561); PdhA, pyruvate dehydrogenase complex E1 component, alpha-subunit (SAOUHSC_01040); PdhB, pyruvate dehydrogenase complex E1 component, beta-subunit (SAOUHSC_01041); PdhC, pyruvate dehydrogenase component E2 (SAOUHSC_01042); CitZ, citrate synthase (SAOUHSC_01802); CitB, aconitate hydratase (SAOUHSC_01347); CitC, isocitrate dehyrogenase (SAOUHSC_01801); OdhA/Kgd, 2-oxoglutarate dehydrogenase E1 component/alpha-ketoglutarate decarboxylase (SAOUHSC_01418); OdhB, 2-oxoglutarate dehydrogenase E2 component/dihydrolipoamide acetyltransferase (SAOUHSC_01416); SucC, succinyl-CoA synthetase subunit beta (SAOUHSC_01216); SucD, succinyl-CoA synthetase subunit alpha (SAOUHSC_01218); SdhC, succinate dehydrogenase cytochrome _b_558 subunit (SAOUHSC_01103); SdhA, succinate dehydrogenase flavoprotein subunit (SAOUHSC_01104); SdhB, succinate dehydrogenase iron-sulfur subunit (SAOUHSC_01105); FumC, fumarate hydratase class II (SAOUHSC_01983); Mqo-2, malate:quinone oxidoreductase (SAOUHSC_02647 and SAOUHSC_02927); Pox, pyruvate oxidase (SAOUHSC_02849); AckA, acetate kinase (SAOUHSC_01820); AcyP, acylphosphatase (SAOUHSC_01406); Pta, phosphate acetyltransferase (SAOUHSC_00574); AcsA, acetyl-CoA synthetase (SAOUHSC_01846).
Similar articles
- Global gene expression in Staphylococcus aureus biofilms.
Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. Beenken KE, et al. J Bacteriol. 2004 Jul;186(14):4665-84. doi: 10.1128/JB.186.14.4665-4684.2004. J Bacteriol. 2004. PMID: 15231800 Free PMC article. - The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase.
Patton TG, Rice KC, Foster MK, Bayles KW. Patton TG, et al. Mol Microbiol. 2005 Jun;56(6):1664-74. doi: 10.1111/j.1365-2958.2005.04653.x. Mol Microbiol. 2005. PMID: 15916614 - Global gene expression in Staphylococcus aureus following exposure to alcohol.
Korem M, Gov Y, Rosenberg M. Korem M, et al. Microb Pathog. 2010 Feb;48(2):74-84. doi: 10.1016/j.micpath.2009.11.002. Epub 2009 Nov 10. Microb Pathog. 2010. PMID: 19900530 - Succinate dehydrogenase: the complex roles of a simple enzyme.
Huang S, Millar AH. Huang S, et al. Curr Opin Plant Biol. 2013 Jun;16(3):344-9. doi: 10.1016/j.pbi.2013.02.007. Epub 2013 Mar 1. Curr Opin Plant Biol. 2013. PMID: 23453781 Review. - Unsuspected task for an old team: succinate, fumarate and other Krebs cycle acids in metabolic remodeling.
Bénit P, Letouzé E, Rak M, Aubry L, Burnichon N, Favier J, Gimenez-Roqueplo AP, Rustin P. Bénit P, et al. Biochim Biophys Acta. 2014 Aug;1837(8):1330-7. doi: 10.1016/j.bbabio.2014.03.013. Epub 2014 Mar 31. Biochim Biophys Acta. 2014. PMID: 24699309 Review.
Cited by
- Phenotype switching is a natural consequence of Staphylococcus aureus replication.
Edwards AM. Edwards AM. J Bacteriol. 2012 Oct;194(19):5404-12. doi: 10.1128/JB.00948-12. Epub 2012 Aug 3. J Bacteriol. 2012. PMID: 22865841 Free PMC article. - Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response.
Painter KL, Strange E, Parkhill J, Bamford KB, Armstrong-James D, Edwards AM. Painter KL, et al. Infect Immun. 2015 May;83(5):1830-44. doi: 10.1128/IAI.03016-14. Epub 2015 Feb 17. Infect Immun. 2015. PMID: 25690100 Free PMC article. - Mechanisms of Bacterial Tolerance and Persistence in the Gastrointestinal and Respiratory Environments.
Trastoy R, Manso T, Fernández-García L, Blasco L, Ambroa A, Pérez Del Molino ML, Bou G, García-Contreras R, Wood TK, Tomás M. Trastoy R, et al. Clin Microbiol Rev. 2018 Aug 1;31(4):e00023-18. doi: 10.1128/CMR.00023-18. Print 2018 Oct. Clin Microbiol Rev. 2018. PMID: 30068737 Free PMC article. Review. - Long-Term Intrahost Evolution of Methicillin Resistant Staphylococcus aureus Among Cystic Fibrosis Patients With Respiratory Carriage.
Azarian T, Ridgway JP, Yin Z, David MZ. Azarian T, et al. Front Genet. 2019 Jun 12;10:546. doi: 10.3389/fgene.2019.00546. eCollection 2019. Front Genet. 2019. PMID: 31244886 Free PMC article. - Modulation of Staphylococcus aureus gene expression during proliferation in platelet concentrates with focus on virulence and platelet functionality.
Yousuf B, Pasha R, Pineault N, Ramirez-Arcos S. Yousuf B, et al. PLoS One. 2024 Jul 25;19(7):e0307920. doi: 10.1371/journal.pone.0307920. eCollection 2024. PLoS One. 2024. PMID: 39052660 Free PMC article.
References
- Archibald, L. K., and R. P. Gaynes. 1997. Hospital-acquired infections in the United States. The importance of interhospital comparisons. Infect. Dis. Clin. North Am. 11:245-255. - PubMed
- Augustin, J., and F. Götz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54:203-207. - PubMed
- Baddour, L. M., and G. D. Christensen. 1987. Prosthetic valve endocarditis due to small-colony staphylococcal variants. Rev. Infect. Dis. 9:1168-1174. - PubMed
- Bates, D. M., C. von Eiff, P. J. McNamara, G. Peters, M. R. Yeaman, A. S. Bayer, and R. A. Proctor. 2003. Staphylococcus aureus menD and hemB mutants are as infective as the parent strains, but the menadione biosynthetic mutant persists within the kidney. J. Infect. Dis. 187:1654-1661. - PubMed
- Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases