Insights into heterocyclization from two highly similar enzymes - PubMed (original) (raw)
Insights into heterocyclization from two highly similar enzymes
John A McIntosh et al. J Am Chem Soc. 2010.
Abstract
The cyanobactin biosynthetic pathways pat and tru, isolated from metagenomes of marine animals, lead to diverse natural products containing heterocycles derived from Cys, Ser, and Thr. Previous work has shown that PatD and TruD are extremely broad-substrate heterocyclase enzymes. These enzymes are virtually identical in their N-terminal putative catalytic domains, but only approximately 77% identical in their C-terminal putative substrate-binding domains. Here, we show that these differences allow the enzymes to control regioselectivity of posttranslational modifications, helping to control product chemistry in this hypervariable family of marine natural products.
Figures
Figure 1
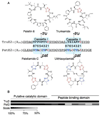
Biosynthesis of heterocyclic cyanobactins. A. Enzymes modify ribosomal precursor peptides into mature products. Trunkamide group: top; patellamide group: bottom. B. An alignment of PatD and TruD proteins, showing putative catalytic and peptide-binding domains. Catalytic domains are >99% identical; peptide-binding domains are >77% identical.
Figure 2
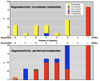
Enzyme selectivity defined by metagenome sequence and chemical analysis. Natural products from the pat (top) and tru (bottom) pathways contain modified Cys, Ser, and Thr residues in the defined positions. y-axis: % of natural products containing these modifications. x-axis: amino acid position in cassette. Empty space above the bars denotes hypervariability: diverse amino acids occupy these positions in isolated natural products.
Figure 3
Reaction characterization and substrate specificity of TruD and PatD. A. MALDI MS of reaction products. B. Position of modifications identified by ESI-MS/MS. C. Representative SDS-PAGE gel showing band shifts of precursor peptides 1–4. The left of each triplet is a standard of the peptide, middle is the TruD-modified peptide, and right is the PatD-modified peptide.
Similar articles
- Marine molecular machines: heterocyclization in cyanobactin biosynthesis.
McIntosh JA, Schmidt EW. McIntosh JA, et al. Chembiochem. 2010 Jul 5;11(10):1413-21. doi: 10.1002/cbic.201000196. Chembiochem. 2010. PMID: 20540059 Free PMC article. - Modularity of RiPP Enzymes Enables Designed Synthesis of Decorated Peptides.
Sardar D, Lin Z, Schmidt EW. Sardar D, et al. Chem Biol. 2015 Jul 23;22(7):907-16. doi: 10.1016/j.chembiol.2015.06.014. Epub 2015 Jul 9. Chem Biol. 2015. PMID: 26165156 Free PMC article. - Recognition sequences and substrate evolution in cyanobactin biosynthesis.
Sardar D, Pierce E, McIntosh JA, Schmidt EW. Sardar D, et al. ACS Synth Biol. 2015 Feb 20;4(2):167-76. doi: 10.1021/sb500019b. Epub 2014 Mar 26. ACS Synth Biol. 2015. PMID: 24625112 Free PMC article. - Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria.
Sivonen K, Leikoski N, Fewer DP, Jokela J. Sivonen K, et al. Appl Microbiol Biotechnol. 2010 May;86(5):1213-25. doi: 10.1007/s00253-010-2482-x. Epub 2010 Feb 27. Appl Microbiol Biotechnol. 2010. PMID: 20195859 Free PMC article. Review. - The structural biology of patellamide biosynthesis.
Koehnke J, Bent AF, Houssen WE, Mann G, Jaspars M, Naismith JH. Koehnke J, et al. Curr Opin Struct Biol. 2014 Dec;29:112-121. doi: 10.1016/j.sbi.2014.10.006. Epub 2014 Nov 25. Curr Opin Struct Biol. 2014. PMID: 25460274 Free PMC article. Review.
Cited by
- Micrococcin cysteine-to-thiazole conversion through transient interactions between the scaffolding protein TclI and the modification enzymes TclJ and TclN.
Calvopina-Chavez DG, Bursey DM, Tseng Y-J, Patil LM, Bewley KD, Bennallack PR, McPhie JM, Wagstaff KB, Daley A, Miller SM, Moody JD, Price JC, Griffitts JS. Calvopina-Chavez DG, et al. Appl Environ Microbiol. 2024 Jun 18;90(6):e0024424. doi: 10.1128/aem.00244-24. Epub 2024 May 23. Appl Environ Microbiol. 2024. PMID: 38780510 Free PMC article. - Non-modular fatty acid synthases yield distinct N-terminal acylation in ribosomal peptides.
Ren H, Huang C, Pan Y, Dommaraju SR, Cui H, Li M, Gadgil MG, Mitchell DA, Zhao H. Ren H, et al. Nat Chem. 2024 Aug;16(8):1320-1329. doi: 10.1038/s41557-024-01491-3. Epub 2024 Mar 25. Nat Chem. 2024. PMID: 38528101 Free PMC article. - Promiscuous Enzymes for Residue-Specific Peptide and Protein Late-Stage Functionalization.
Alexander AK, Elshahawi SI. Alexander AK, et al. Chembiochem. 2023 Sep 1;24(17):e202300372. doi: 10.1002/cbic.202300372. Epub 2023 Jul 19. Chembiochem. 2023. PMID: 37338668 Free PMC article. Review. - Chemoenzymatic Late-Stage Modifications Enable Downstream Click-Mediated Fluorescent Tagging of Peptides.
Colombano A, Dalponte L, Dall'Angelo S, Clemente C, Idress M, Ghazal A, Houssen WE. Colombano A, et al. Angew Chem Int Ed Engl. 2023 Apr 11;62(16):e202215979. doi: 10.1002/anie.202215979. Epub 2023 Mar 10. Angew Chem Int Ed Engl. 2023. PMID: 36815722 Free PMC article. - Biochemical characterization of a cyanobactin arginine-_N_-prenylase from the autumnalamide biosynthetic pathway.
Clemente C, Johnson N, Ouyang X, Popin RV, Dall'Angelo S, Wahlsten M, Jokela J, Colombano A, Nardone B, Fewer DP, Houssen WE. Clemente C, et al. Chem Commun (Camb). 2022 Oct 27;58(86):12054-12057. doi: 10.1039/d2cc01799g. Chem Commun (Camb). 2022. PMID: 36193595 Free PMC article.
References
- Schloss PD, Handelsman J. Curr. Opin. Biotechnol. 2003;14:303–310. - PubMed
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Chem. Biol. 1998;5:R245–R249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM071425/GM/NIGMS NIH HHS/United States
- R01 GM071425-05/GM/NIGMS NIH HHS/United States
- R01 GM071425/GM/NIGMS NIH HHS/United States
- R01 GM071425-01A1/GM/NIGMS NIH HHS/United States
- R01 GM071425-04/GM/NIGMS NIH HHS/United States
- R01 GM071425-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases