Receiver domain structure and function in response regulator proteins - PubMed (original) (raw)
Review
Receiver domain structure and function in response regulator proteins
Robert B Bourret. Curr Opin Microbiol. 2010 Apr.
Abstract
During signal transduction by two-component regulatory systems, sensor kinases detect and encode input information while response regulators (RRs) control output. Most receiver domains function as phosphorylation-mediated switches within RRs, but some transfer phosphoryl groups in multistep phosphorelays. Conserved features of receiver domain amino acid sequence correlate with structure and hence function. Receiver domains catalyze their own phosphorylation and dephosphorylation in reactions requiring a divalent cation. Molecular dynamics simulations are supplementing structural investigation of the conformational changes that underlie receiver domain switch function. As understanding of features shared by all receiver domains matures, factors conferring differences (e.g. in reaction rate or specificity) are receiving increased attention. Numerous examples of atypical receiver or pseudo-receiver domains that function without phosphorylation have recently been characterized.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
Figure 1
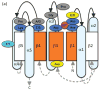
Schematic diagram of the relationship between receiver domain amino acid sequence and basic structural elements as described in the text, with the active site viewed from the (a) side or (b) top. Five α-helices surround a parallel five-stranded β-sheet. Loops connecting strands and helices are shown as black solid lines on the active site side of the domain and gray dashed lines on the opposite side, with arrowheads indicating N- to C-terminal direction. Orange indicates pattern of conserved hydrophobic residues on the central β-strands and faces of three α-helices. The highly conserved residues of the active site quintet are in blue, with the moderately conserved aromatic residue in cyan. Divalent metal ion is magenta. Residues presumed to be strongly conserved for structural reasons are in gray. Frequently conserved acidic residues of unknown function are in yellow.
Figure 1
Schematic diagram of the relationship between receiver domain amino acid sequence and basic structural elements as described in the text, with the active site viewed from the (a) side or (b) top. Five α-helices surround a parallel five-stranded β-sheet. Loops connecting strands and helices are shown as black solid lines on the active site side of the domain and gray dashed lines on the opposite side, with arrowheads indicating N- to C-terminal direction. Orange indicates pattern of conserved hydrophobic residues on the central β-strands and faces of three α-helices. The highly conserved residues of the active site quintet are in blue, with the moderately conserved aromatic residue in cyan. Divalent metal ion is magenta. Residues presumed to be strongly conserved for structural reasons are in gray. Frequently conserved acidic residues of unknown function are in yellow.
Figure 2
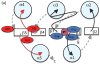
Schematic diagram indicating key differences between (a) inactive and (b) active conformations. View and color coding as in Figure 1b, with some features removed for clarity. Residues and loops that undergo the most significant changes are shown in red (inactive) or dark green (active). Phosphoryl group and lines indicating key hydrogen bonds are in light green.
Figure 2
Schematic diagram indicating key differences between (a) inactive and (b) active conformations. View and color coding as in Figure 1b, with some features removed for clarity. Residues and loops that undergo the most significant changes are shown in red (inactive) or dark green (active). Phosphoryl group and lines indicating key hydrogen bonds are in light green.
Figure 3
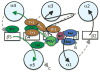
Variable active site residues affect autodephosphorylation kinetics. View and color coding as in Figure 2b. The amino acids located one and two positions to the C-terminal sides of the Asp phosphorylation site and the highly conserved Thr/Ser are shown in brown. Established and putative roles are described in the text.
Similar articles
- Phyletic Distribution and Lineage-Specific Domain Architectures of Archaeal Two-Component Signal Transduction Systems.
Galperin MY, Makarova KS, Wolf YI, Koonin EV. Galperin MY, et al. J Bacteriol. 2018 Mar 12;200(7):e00681-17. doi: 10.1128/JB.00681-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29263101 Free PMC article. - Proposed Role for KaiC-Like ATPases as Major Signal Transduction Hubs in Archaea.
Makarova KS, Galperin MY, Koonin EV. Makarova KS, et al. mBio. 2017 Dec 5;8(6):e01959-17. doi: 10.1128/mBio.01959-17. mBio. 2017. PMID: 29208747 Free PMC article. - The many faces of the helix-turn-helix domain: transcription regulation and beyond.
Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. Aravind L, et al. FEMS Microbiol Rev. 2005 Apr;29(2):231-62. doi: 10.1016/j.femsre.2004.12.008. FEMS Microbiol Rev. 2005. PMID: 15808743 Review. - Use of restrained molecular dynamics to predict the conformations of phosphorylated receiver domains in two-component signaling systems.
Foster CA, West AH. Foster CA, et al. Proteins. 2017 Jan;85(1):155-176. doi: 10.1002/prot.25207. Epub 2016 Nov 20. Proteins. 2017. PMID: 27802580 Free PMC article. - Diversity of structure and function of response regulator output domains.
Galperin MY. Galperin MY. Curr Opin Microbiol. 2010 Apr;13(2):150-9. doi: 10.1016/j.mib.2010.01.005. Epub 2010 Mar 11. Curr Opin Microbiol. 2010. PMID: 20226724 Free PMC article. Review.
Cited by
- Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic Di-GMP signaling.
Chen MW, Kotaka M, Vonrhein C, Bricogne G, Rao F, Chuah ML, Svergun D, Schneider G, Liang ZX, Lescar J. Chen MW, et al. J Bacteriol. 2012 Sep;194(18):4837-46. doi: 10.1128/JB.00560-12. Epub 2012 Jun 29. J Bacteriol. 2012. PMID: 22753070 Free PMC article. - Mapping allosteric communications within individual proteins.
Wang J, Jain A, McDonald LR, Gambogi C, Lee AL, Dokholyan NV. Wang J, et al. Nat Commun. 2020 Jul 31;11(1):3862. doi: 10.1038/s41467-020-17618-2. Nat Commun. 2020. PMID: 32737291 Free PMC article. - Promoter recognition and activation by the global response regulator CbrB in Pseudomonas aeruginosa.
Abdou L, Chou HT, Haas D, Lu CD. Abdou L, et al. J Bacteriol. 2011 Jun;193(11):2784-92. doi: 10.1128/JB.00164-11. Epub 2011 Apr 8. J Bacteriol. 2011. PMID: 21478360 Free PMC article. - Francisella novicida Two-Component System Response Regulator BfpR Modulates iglC Gene Expression, Antimicrobial Peptide Resistance, and Biofilm Production.
Dean SN, Milton ME, Cavanagh J, van Hoek ML. Dean SN, et al. Front Cell Infect Microbiol. 2020 Mar 13;10:82. doi: 10.3389/fcimb.2020.00082. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32232010 Free PMC article. - The residue threonine 82 of DevR (DosR) is essential for DevR activation and function in Mycobacterium tuberculosis despite its atypical location.
Gautam US, Sikri K, Tyagi JS. Gautam US, et al. J Bacteriol. 2011 Sep;193(18):4849-58. doi: 10.1128/JB.05051-11. Epub 2011 Jul 15. J Bacteriol. 2011. PMID: 21764934 Free PMC article.
References
- Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials