Emerging regulatory paradigms for control of gene expression by 1,25-dihydroxyvitamin D3 - PubMed (original) (raw)
Emerging regulatory paradigms for control of gene expression by 1,25-dihydroxyvitamin D3
J Wesley Pike et al. J Steroid Biochem Mol Biol. 2010 Jul.
Abstract
1,25-dihydroxyvitamin D3 (1,25(OH)2D3) functions as a steroid hormone to modulate the expression of genes. Its actions are mediated by the vitamin D receptor (VDR) which binds to target genes and functions to recruit coregulatory complexes that are essential for transcriptional modulation. ChIP analysis coupled to tiled DNA microarray hybridization (ChIP-chip) or massively parallel DNA sequencing (ChIP-seq) is now providing critical new insight into how genes are regulated. In studies herein, we utilized these techniques as well as gene expression analysis to explore the actions of 1,25(OH)2D3 at the genome-wide and individual target gene levels in cells. We identify a series of overarching principles that likely define the actions of 1,25(OH)2D3 at most target genes. We discover that while VDR binding to target sites is ligand-dependent, RXR binding is ligand-independent. We also show that while VDR/RXR binding can localize to promoters, it occurs more frequently at multiple sites many kilobases from target gene promoters. We then describe a new method whereby the regulatory regions of complex genes can be evaluated using large recombineered bacterial artificial chromosomes. We conclude that these new approaches are likely to replace many of the traditional methods used to explore the regulation of transcription.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures
Figure 1
ChIP-chip analysis of basal and 1,25(OH)2D3-induced VDR and RXR binding to the mouse Vdr gene. MC3T3-E1 cells were treated with either vehicle or 1,25(OH)2D3 for 3 hr and then subjected to ChIP-chip analysis using antibodies to VDR or RXR. A schematic diagram of the mouse Vdr gene locus and its location on chromosome 15 is depicted at the top. Previously identified enhancers are designated S1/S2, S3, PP and U1 and marked through vertical shading. VDR data tracks represent the log2 ratios of fluorescence obtained from vehicle- or 1,25(OH)2D3-treated samples precipitated with antibody to the VDR vs corresponding sample input DNAs. RXR data tracks correspond to equivalent analyses. Red peaks represent statistically valid regions of VDR or RXR binding (FDR <0.05).
Figure 2
ChIP-chip analysis of basal and 1,25(OH)2D3 induced RNA polymerase II density and histone 4 acetylation analysis to the mouse Vdr gene. The schematic at the top of the figure is described in Figure 1. RNA polymerase II data tracks (RNA pol II) represent the log2 ratios of fluorescence obtained from vehicle- or 1,25(OH)2D3-treated samples precipitated with antibody to the RNA polymerase II vs corresponding sample input DNA’s. Histone H4 acetylation data tracks (H4-Ac) correspond to equivalent analyses. Red peaks represent statistically valid regions of VDR or RXR binding (FDR <0.05).
Figure 3
Summary model documenting the interaction of transcription factors at the mouse VDR gene locus. MC3T3-E1 cells were treated with either vehicle, 1,25(OH)2D3, retinoic acid, dexamethasone or forskolin for 6 hrs and then subjected to ChIP-chip analysis using antibodies to VDR, RXR, RAR, GR, GREB or C/EBP□. Positive binding activity in the presence of the appropriate activating ligand is indicated by a colored circle at enhancers identified for the mouse Vdr gene.
Figure 4
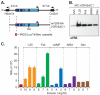
BAC clone analysis of the mouse Vdr gene locus in stably transfected MC3T3-E1 cells. Cells were stably transfected with the recombinant Vdr BAC clone depicted in (A). B, Western blot analysis of recombinant VDR expression in MC3T3-E1 cells stably transfected with the recombinant Vdr BAC clone depicted in (A) and its induction by 1,25(OH)2D3, RA, Dex, forskolin. C, Analysis of luciferase activity in MC3T3-E1 cells stably transfected with the recombinant Vdr BAC clone depicted in (A) and its dose dependent induction by 1,25(OH)2D3, RA, Dex and forskolin. Data represent the average of a triplicate set of reactions ± SEM.
Similar articles
- Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3.
Zella LA, Kim S, Shevde NK, Pike JW. Zella LA, et al. Mol Endocrinol. 2006 Jun;20(6):1231-47. doi: 10.1210/me.2006-0015. Epub 2006 Feb 23. Mol Endocrinol. 2006. PMID: 16497728 - Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone.
Meyer MB, Goetsch PD, Pike JW. Meyer MB, et al. J Steroid Biochem Mol Biol. 2010 Jul;121(1-2):136-41. doi: 10.1016/j.jsbmb.2010.02.011. Epub 2010 Feb 18. J Steroid Biochem Mol Biol. 2010. PMID: 20171278 Free PMC article. - The vitamin D hormone and its nuclear receptor: molecular actions and disease states.
Haussler MR, Haussler CA, Jurutka PW, Thompson PD, Hsieh JC, Remus LS, Selznick SH, Whitfield GK. Haussler MR, et al. J Endocrinol. 1997 Sep;154 Suppl:S57-73. J Endocrinol. 1997. PMID: 9379138 Review. - The parathyroid hormone-regulated transcriptome in osteocytes: parallel actions with 1,25-dihydroxyvitamin D3 to oppose gene expression changes during differentiation and to promote mature cell function.
St John HC, Meyer MB, Benkusky NA, Carlson AH, Prideaux M, Bonewald LF, Pike JW. St John HC, et al. Bone. 2015 Mar;72:81-91. doi: 10.1016/j.bone.2014.11.010. Epub 2014 Nov 22. Bone. 2015. PMID: 25460572 Free PMC article. - What do we learn from the genome-wide perspective on vitamin D3?
Carlberg C. Carlberg C. Anticancer Res. 2015 Feb;35(2):1143-51. Anticancer Res. 2015. PMID: 25667505 Review.
Cited by
- Vitamin D and cancer: a review of molecular mechanisms.
Fleet JC, DeSmet M, Johnson R, Li Y. Fleet JC, et al. Biochem J. 2012 Jan 1;441(1):61-76. doi: 10.1042/BJ20110744. Biochem J. 2012. PMID: 22168439 Free PMC article. Review. - A brief review of vitamin D as a potential target for the regulation of blood glucose and inflammation in diabetes-associated periodontitis.
Dong C, Hu X, Tripathi AS. Dong C, et al. Mol Cell Biochem. 2022 Sep;477(9):2257-2268. doi: 10.1007/s11010-022-04445-w. Epub 2022 Apr 27. Mol Cell Biochem. 2022. PMID: 35478388 Review. - Ileal Transcriptome Profiles of Japanese Quail Divergent in Phosphorus Utilization.
Oster M, Reyer H, Trakooljul N, Weber FM, Xi L, Muráni E, Ponsuksili S, Rodehutscord M, Bennewitz J, Wimmers K. Oster M, et al. Int J Mol Sci. 2020 Apr 16;21(8):2762. doi: 10.3390/ijms21082762. Int J Mol Sci. 2020. PMID: 32316159 Free PMC article. - Curcumin induces human cathelicidin antimicrobial peptide gene expression through a vitamin D receptor-independent pathway.
Guo C, Rosoha E, Lowry MB, Borregaard N, Gombart AF. Guo C, et al. J Nutr Biochem. 2013 May;24(5):754-9. doi: 10.1016/j.jnutbio.2012.04.002. Epub 2012 Jul 25. J Nutr Biochem. 2013. PMID: 22841393 Free PMC article. - Targeting CSC-related miRNAs for cancer therapy by natural agents.
Bao B, Li Y, Ahmad A, Azmi AS, Bao G, Ali S, Banerjee S, Kong D, Sarkar FH. Bao B, et al. Curr Drug Targets. 2012 Dec;13(14):1858-68. doi: 10.2174/138945012804545515. Curr Drug Targets. 2012. PMID: 23140295 Free PMC article. Review.
References
- DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80(6 Suppl):1689S–1696S. - PubMed
- Jurutka PW, Whitfield GK, Hsieh JC, Thompson PD, Haussler CA, Haussler MR. Molecular nature of the vitamin D receptor and its role in regulation of gene expression. Rev Endocr Metab Disord. 2001;2(2):203–216. - PubMed
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95(7):927–937. - PubMed
- Vanhooke J, Benning M, Bauer C, Pike J, DeLuca H. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. 2004;43(14):4101–4110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK-074993/DK/NIDDK NIH HHS/United States
- R01 DK072281-04/DK/NIDDK NIH HHS/United States
- R01 DK072281/DK/NIDDK NIH HHS/United States
- R01 DK073995/DK/NIDDK NIH HHS/United States
- R01 DK074993-03S1/DK/NIDDK NIH HHS/United States
- R01 DK072281-05/DK/NIDDK NIH HHS/United States
- R01 DK074993/DK/NIDDK NIH HHS/United States
- DK-072281/DK/NIDDK NIH HHS/United States
- R01 DK073995-02/DK/NIDDK NIH HHS/United States
- R01 DK074993-03/DK/NIDDK NIH HHS/United States
- AR-045173/AR/NIAMS NIH HHS/United States
- R01 AR045173-05/AR/NIAMS NIH HHS/United States
- R01 AR045173/AR/NIAMS NIH HHS/United States
- R01 DK074993-04/DK/NIDDK NIH HHS/United States
- DK-073995/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources