Anaerobic sulfatase-maturating enzyme--a mechanistic link with glycyl radical-activating enzymes? - PubMed (original) (raw)
Anaerobic sulfatase-maturating enzyme--a mechanistic link with glycyl radical-activating enzymes?
Alhosna Benjdia et al. FEBS J. 2010 Apr.
Abstract
Sulfatases form a major group of enzymes present in prokaryotes and eukaryotes. This class of hydrolases is unique in requiring essential post-translational modification of a critical active-site cysteinyl or seryl residue to C(alpha)-formylglycine (FGly). Herein, we report mechanistic investigations of a unique class of radical-S-adenosyl-L-methionine (AdoMet) enzymes, namely anaerobic sulfatase-maturating enzymes (anSMEs), which catalyze the oxidation of Cys-type and Ser-type sulfatases and possess three [4Fe-4S](2+,+) clusters. We were able to develop a reliable quantitative enzymatic assay that allowed the direct measurement of FGly production and AdoMet cleavage. The results demonstrate stoichiometric coupling of AdoMet cleavage and FGly formation using peptide substrates with cysteinyl or seryl active-site residues. Analytical and EPR studies of the reconstituted wild-type enzyme and cysteinyl cluster mutants indicate the presence of three almost isopotential [4Fe-4S](2+,+) clusters, each of which is required for the generation of FGly in vitro. More surprisingly, our data indicate that the two additional [4Fe-4S](2+,+) clusters are required to obtain efficient reductive cleavage of AdoMet, suggesting their involvement in the reduction of the radical AdoMet [4Fe-4S](2+,+) center. These results, in addition to the recent demonstration of direct abstraction by anSMEs of the C(beta) H-atom from the sulfatase active-site cysteinyl or seryl residue using a 5'-deoxyadenosyl radical, provide new insights into the mechanism of this new class of radical-AdoMet enzymes.
Figures
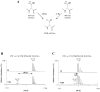
Figure 1. Sulfatase maturation scheme leading from a cysteinyl or seryl residue to a Cα-formylglycine residue (FGly) in sulfatase active site (A) – MALDI-TOF MS analysis of maturation of peptide 17C (B) and 17S (C) incubated 2 and 12 hours with anSMEcpe respectively
AnSMEcpe was incubated with each peptide (500 μM) under reducing conditions in the presence of AdoMet (1 mM).
Figure 2. HPLC analysis of incubation reactions with peptide 17C (A) or 17S (B) and time-dependant formation of FGly-containing peptide (C) and 5′-deoxyadenosine (D) by anSMEcpe
AnSMEcpe was incubated with 17C (◆), 17S (▲) or 17A (●) peptide (500 μM) under reducing conditions in the presence of AdoMet (1 mM), DTT (6 mM) and dithionite (3 mM).
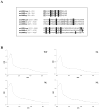
Figure 3. Sequence alignment of the three anSMEs putative clusters
(A) anSMEcpe (CPF_0616 from Clostridium perfringens), anSMEbt (BT_0238 from Bacteroides thetaiotaomicron) and anSMEkp (AtsB from Klebsiella pneumoniae). Sequence positions in the proteins are in brackets. The conserved cysteinyl residues are indicated in black boxes and the other conserved residues are shadowed – UV visible absorption specta of reconstituted wild-type (WT) and M1, M2 and M3 variants of anSMEbt (B).
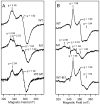
Figure 4. X-band EPR spectra of dithionite-reduced reconstituted samples of WT and M1 mutant anSMEbt in the absence (A) and presence (B) of a 20-fold stoichiometric excess of AdoMet
The WT minus M1 mutant difference spectrum at the bottom of each panel corresponds to the EPR spectrum of the S = 1/2 [4Fe-4S]+ radical-AdoMet cluster with (B) and without (A) AdoMet bound at the unique Fe site. EPR spectra were recorded at 10 K with 20 mW microwave power, 0.65 mT modulation amplitude and a microwave frequency of 9.603 GHz. The spectrometer gain was 2-fold higher for the samples prepared without AdoMet. Samples of WT and M1 mutant anSMEbt (each 0.4 mM) in Tris-HCl buffer, pH 7.5, were anaerobically reduced with a 10-fold stoichiometric excess of sodium dithionite.
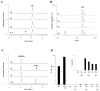
Figure 5. HPLC (A) and MALDI-TOF MS (B) analysis of the peptide maturation catalyzed by WT and M1, M2 and M3 mutants of anSMEbt
The WT and mutant forms of anSMEbt (each 60 μM) were incubated with 17C peptide (500 μM) under reducing conditions in the presence of AdoMet (1 mM), DTT (6 mM) and dithionite (3 mM) for 4 hours under anaerobic and reducing conditions. (C) HPLC analysis of AdoMet cleavage catalyzed by WT or M1, M2 and M3 mutants of anSMEbt in presence of 17C peptide. (D) Relative production of 5′-dA compared to the WT enzyme with or without substrate peptide (inset: magnified picture of the results obtained for the mutants).
Figure 6. Sequence alignment of anSMEcpe, QHNDH-AE, PqqE and the ST protein
Sequence positions in the proteins are in brackets. The percentage of similarity between the corresponding region of anSME and the different enzymes is indicated.
Figure 7. Two possible mechanisms for anSMEs with Cys-type sulfatases substrates
(A) After reduction of the radical AdoMet [4Fe-4S] center, AdoMet is reductively cleaved and the resulting 5′-deoxyadenosyl radical abstracts a Cβ hydrogen atom from cysteinyl residue of the substrate peptide that is ligated to a unique site of a [4Fe-4S] center (Cluster II). Cluster III is proposed to play a role in mediating electron transfer from the physiological electron to the radical AdoMet [4Fe-4S] cluster or from Cluster II to the physiological electron acceptor. (B) The peptidyl substrate is first deprotonated and AdoMet is reductively cleaved. The resulting 5′-deoxyadenosyl radical abstracts a Cβ hydrogen atom from cysteinyl residue to generate a substrate radical that is converted to the thioaldehyde intermediate via outer-sphere electron transfer to the radical AdoMet cluster. In this scheme, the additional [4Fe-4S] centers, Clusters II and III, have a key role in mediating the initial electron transfer from the physiological electron to the radical AdoMet [4Fe-4S] cluster. In both mechanisms, a thioaldehyde intermediate is formed and further hydrolyzed to form the FGly residue with the release of hydrogen disulfide.
Figure 7. Two possible mechanisms for anSMEs with Cys-type sulfatases substrates
(A) After reduction of the radical AdoMet [4Fe-4S] center, AdoMet is reductively cleaved and the resulting 5′-deoxyadenosyl radical abstracts a Cβ hydrogen atom from cysteinyl residue of the substrate peptide that is ligated to a unique site of a [4Fe-4S] center (Cluster II). Cluster III is proposed to play a role in mediating electron transfer from the physiological electron to the radical AdoMet [4Fe-4S] cluster or from Cluster II to the physiological electron acceptor. (B) The peptidyl substrate is first deprotonated and AdoMet is reductively cleaved. The resulting 5′-deoxyadenosyl radical abstracts a Cβ hydrogen atom from cysteinyl residue to generate a substrate radical that is converted to the thioaldehyde intermediate via outer-sphere electron transfer to the radical AdoMet cluster. In this scheme, the additional [4Fe-4S] centers, Clusters II and III, have a key role in mediating the initial electron transfer from the physiological electron to the radical AdoMet [4Fe-4S] cluster. In both mechanisms, a thioaldehyde intermediate is formed and further hydrolyzed to form the FGly residue with the release of hydrogen disulfide.
Similar articles
- Mechanistic investigations of anaerobic sulfatase-maturating enzyme: direct Cbeta H-atom abstraction catalyzed by a radical AdoMet enzyme.
Benjdia A, Leprince J, Sandström C, Vaudry H, Berteau O. Benjdia A, et al. J Am Chem Soc. 2009 Jun 24;131(24):8348-9. doi: 10.1021/ja901571p. J Am Chem Soc. 2009. PMID: 19489556 - Anaerobic sulfatase-maturating enzymes, first dual substrate radical S-adenosylmethionine enzymes.
Benjdia A, Subramanian S, Leprince J, Vaudry H, Johnson MK, Berteau O. Benjdia A, et al. J Biol Chem. 2008 Jun 27;283(26):17815-26. doi: 10.1074/jbc.M710074200. Epub 2008 Apr 11. J Biol Chem. 2008. PMID: 18408004 Free PMC article. - Further characterization of Cys-type and Ser-type anaerobic sulfatase maturating enzymes suggests a commonality in the mechanism of catalysis.
Grove TL, Ahlum JH, Qin RM, Lanz ND, Radle MI, Krebs C, Booker SJ. Grove TL, et al. Biochemistry. 2013 Apr 30;52(17):2874-87. doi: 10.1021/bi400136u. Epub 2013 Apr 16. Biochemistry. 2013. PMID: 23477283 Free PMC article. - Pyruvate formate-lyase activating enzyme: elucidation of a novel mechanism for glycyl radical formation.
Buis JM, Broderick JB. Buis JM, et al. Arch Biochem Biophys. 2005 Jan 1;433(1):288-96. doi: 10.1016/j.abb.2004.09.028. Arch Biochem Biophys. 2005. PMID: 15581584 Review. - Formylglycine, a post-translationally generated residue with unique catalytic capabilities and biotechnology applications.
Appel MJ, Bertozzi CR. Appel MJ, et al. ACS Chem Biol. 2015 Jan 16;10(1):72-84. doi: 10.1021/cb500897w. ACS Chem Biol. 2015. PMID: 25514000 Free PMC article. Review.
Cited by
- Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A. Kubiak X, et al. Nat Chem Biol. 2024 Mar;20(3):382-391. doi: 10.1038/s41589-023-01493-1. Epub 2023 Dec 29. Nat Chem Biol. 2024. PMID: 38158457 - Emerging themes in radical SAM chemistry.
Shisler KA, Broderick JB. Shisler KA, et al. Curr Opin Struct Biol. 2012 Dec;22(6):701-10. doi: 10.1016/j.sbi.2012.10.005. Epub 2012 Nov 8. Curr Opin Struct Biol. 2012. PMID: 23141873 Free PMC article. Review. - Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron.
Benjdia A, Martens EC, Gordon JI, Berteau O. Benjdia A, et al. J Biol Chem. 2011 Jul 22;286(29):25973-82. doi: 10.1074/jbc.M111.228841. Epub 2011 Apr 20. J Biol Chem. 2011. PMID: 21507958 Free PMC article. - Radical SAM Enzymes and Ribosomally-Synthesized and Post-translationally Modified Peptides: A Growing Importance in the Microbiomes.
Benjdia A, Berteau O. Benjdia A, et al. Front Chem. 2021 Jul 19;9:678068. doi: 10.3389/fchem.2021.678068. eCollection 2021. Front Chem. 2021. PMID: 34350157 Free PMC article. Review.
References
- Muller I, Kahnert A, Pape T, Sheldrick GM, Meyer-Klaucke W, Dierks T, Kertesz M, Uson I. Crystal structure of the alkylsulfatase AtsK: insights into the catalytic mechanism of the Fe(II) alpha-ketoglutarate-dependent dioxygenase superfamily. Biochemistry. 2004;43:3075–3088. - PubMed
- Hanson SR, Best MD, Wong CH. Sulfatases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl. 2004;43:5736–5763. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM062524/GM/NIGMS NIH HHS/United States
- R01 GM062524-09/GM/NIGMS NIH HHS/United States
- R37 GM062524/GM/NIGMS NIH HHS/United States
- GM62524/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources