Improvement of tissue preparation for laser capture microdissection: application for cell type-specific miRNA expression profiling in colorectal tumors - PubMed (original) (raw)
Improvement of tissue preparation for laser capture microdissection: application for cell type-specific miRNA expression profiling in colorectal tumors
Shuyang Wang et al. BMC Genomics. 2010.
Abstract
Background: Laser capture microdissection (LCM) has successfully isolated pure cell populations from tissue sections and the combination of LCM with standard genomic and proteomic methods has revolutionized molecular analysis of complex tissue. However, the quantity and quality of material recovered after LCM is often still limited for analysis by using whole genomic and proteomic approaches. To procure high quality and quantity of RNA after LCM, we optimized the procedures on tissue preparations and applied the approach for cell type-specific miRNA expression profiling in colorectal tumors.
Results: We found that the ethanol fixation of tissue sections for 2 hours had the maximum improvement of RNA quality (1.8 fold, p = 0.0014) and quantity (1.5 fold, p = 0.066). Overall, the quality (RNA integrity number, RIN) for the microdissected colorectal tissues was 5.2 +/- 1.5 (average +/- SD) for normal (n = 43), 5.7 +/- 1.1 for adenomas (n = 14) and 7.2 +/- 1.2 for carcinomas (n = 44). We then compared miRNA expression profiles of 18 colorectal tissues (6 normal, 6 adenomas and 6 carcinomas) between LCM selected epithelial cells versus stromal cells using Agilent miRNA microarrays. We identified 51 differentially expressed miRNAs (p <= 0.001) between these two cell types. We found that the miRNAs in the epithelial cells could differentiate adenomas from normal and carcinomas. However, the miRNAs in the stromal and mixed cells could not separate adenomas from normal tissues. Finally, we applied quantitative RT-PCR to cross-verify the expression patterns of 7 different miRNAs using 8 LCM-selected epithelial cells and found the excellent correlation of the fold changes between the two platforms (R = 0.996).
Conclusions: Our study demonstrates the feasibility and potential power of discovering cell type-specific miRNA biomarkers in complex tissue using combination of LCM with genome-wide miRNA analysis.
Figures
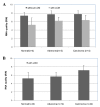
Figure 1
Effect of ethanol fixation on RNA quality and quantity. A) RNA quality (RIN scores) of the tissue sections in the presence (n = 24) and absence (n = 24) of ethanol fixation; B) RNA quantity (ng) of the tissue sections in the presence (n = 24) and absence (n = 24) of ethanol fixation; C) RIN scores of the tissue sections over four time points in the presence (n = 6 per time point) and absence (n = 6 per time point) of ethanol fixation and D) RNA quantity (ng) of the tissue sections over four time points in the presence (n = 6 per time point) and absence (n = 6 per time point) of ethanol fixation. Error bars indicate the corresponding SD. The large errors of the experiments were due to the fact that each tested group consisted of three different tissue types (normal, adenoma and carcinoma) which had the different RNA quality and quantity.
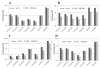
Figure 2
Effect of LCM on RNA quality. A) RNA quality (RIN scores) of the hematoxylin-stained sections with (n = 11) and without (n = 11) LCM and B) RNA quality (RIN scores) of the LCM selected epithelial cells derived from 43 normal, 14 adenoma and 44 carcinoma tissues. Error bars indicate the corresponding SD.
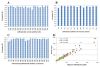
Figure 3
Reliability of LCM and miRNA analysis. A) correlation amongst individual samples of epithelial cells derived from 24 normal colorectal tissues; B) correlation amongst individual samples of epithelial cells derived from 13 colorectal tubular adenomas; C) correlation amongst individual samples of epithelial cells derived from 24 colorectal Dukes' C carcinomas and D) correlation amongst triplicate LCM experiments.
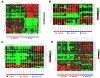
Figure 4
Cell type-specific miRNA expression profiles. A) hierarchical clustering of 51 miRNA expression profiles in LCM selected epithelial and stromal cells from 18 colorectal tissues (n = 6 normal, n = 6 adenomas and n = 6 carcinomas); B) hierarchical clustering of 26 miRNA expression profiles in LCM selected epithelial cells from the colorectal tissues; C) hierarchical clustering of 21 miRNA expression profiles in LCM selected stromal cells from the colorectal tissues and D) hierarchical clustering of 46 miRNA expression profiles in the mixed cell types (epithelial and stromal cells) from the colorectal tissues. The mean signal from biological replicate samples was used for the clustering. Colored bars indicate the range of normalized log2-based signals.
Figure 5
Across-platform comparison. A) comparison of the fold changes in sample pair 54 determined by Agilent miRNA microarrays and by quantitative RT-PCR (54AL: LCM-selected epithelial cells of normal colorectal tissue; 54BL: LCM-selected epithelial cells of Dukes' B carcinomas); B) comparison of the fold changes in sample pair 62 determined by Agilent miRNA microarrays and by quantitative RT-PCR (62AL: LCM-selected epithelial cells of normal colorectal tissue; 62BL: LCM-selected epithelial cells of Dukes' B carcinoma); C) comparison of the fold changes in sample pair 63 determined by Agilent miRNA microarrays and by quantitative RT-PCR (63AL: LCM-selected epithelial cells of normal colorectal tissue; 63BL: LCM-selected epithelial cells of Dukes' C carcinoma) and D) comparison of the fold changes in the sample pair 65 determined by Agilent miRNA microarrays and by quantitative RT-PCR (65AL: LCM-selected epithelial cells of normal colorectal tissue; 65BL: LCM-selected epithelial cells of Dukes' D carcinoma). R indicates the average correlation of 7 individual miRNAs.
Figure 6
Laser capture microdissection of colorectal cells. A) normal; B) adenoma and C) carcinoma. 1) H&E-stained slide (× 20); 2) hematoxylin stained slide before LCM (× 20); 3) hematoxylin stained slide after LCM (× 20) and 4) cap showing adherent cells (× 20).
Similar articles
- Optimized procedures for microarray analysis of histological specimens processed by laser capture microdissection.
Upson JJ, Stoyanova R, Cooper HS, Patriotis C, Ross EA, Boman B, Clapper ML, Knudson AG, Bellacosa A. Upson JJ, et al. J Cell Physiol. 2004 Dec;201(3):366-73. doi: 10.1002/jcp.20073. J Cell Physiol. 2004. PMID: 15389559 - Comparison of progestin transcriptional profiles in rat mammary gland using Laser Capture Microdissection and whole tissue-sampling.
Mazurek N, Frisk AL, Beekman JM, Hartwig A, Meyer K. Mazurek N, et al. Exp Toxicol Pathol. 2013 Nov;65(7-8):949-60. doi: 10.1016/j.etp.2013.01.009. Epub 2013 Mar 7. Exp Toxicol Pathol. 2013. PMID: 23466250 - Assessment of gene expression in head and neck carcinoma using laser capture microdissection and real-time reverse transcription polymerase chain reaction.
Malhotra PS, Malekfzali A, Bonner RF, Juhn S, Van Waes C, Chen Z. Malhotra PS, et al. Laryngoscope. 2004 Dec;114(12):2123-8. doi: 10.1097/01.mlg.0000149446.14770.52. Laryngoscope. 2004. PMID: 15564832 - Laser-controlled microdissection of tissues opens a window of new opportunities.
Hergenhahn M, Kenzelmann M, Gröne HJ. Hergenhahn M, et al. Pathol Res Pract. 2003;199(6):419-23. doi: 10.1078/0344-0338-00440. Pathol Res Pract. 2003. PMID: 12924444 Review. - Application of laser-capture microdissection to analysis of gene expression in the testis.
Sluka P, O'Donnell L, McLachlan RI, Stanton PG. Sluka P, et al. Prog Histochem Cytochem. 2008;42(4):173-201. doi: 10.1016/j.proghi.2007.10.001. Prog Histochem Cytochem. 2008. PMID: 18243898 Review.
Cited by
- Skin Diseases in Laboratory Mice: Approaches to Drug Target Identification and Efficacy Screening.
Sundberg JP, Silva KA, King LE Jr, Pratt CH. Sundberg JP, et al. Methods Mol Biol. 2016;1438:199-224. doi: 10.1007/978-1-4939-3661-8_12. Methods Mol Biol. 2016. PMID: 27150092 Free PMC article. - Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations.
Kadakkuzha BM, Liu XA, McCrate J, Shankar G, Rizzo V, Afinogenova A, Young B, Fallahi M, Carvalloza AC, Raveendra B, Puthanveettil SV. Kadakkuzha BM, et al. Front Cell Neurosci. 2015 Mar 6;9:63. doi: 10.3389/fncel.2015.00063. eCollection 2015. Front Cell Neurosci. 2015. PMID: 25798087 Free PMC article. - MicroRNA profiling in prostate cancer--the diagnostic potential of urinary miR-205 and miR-214.
Srivastava A, Goldberger H, Dimtchev A, Ramalinga M, Chijioke J, Marian C, Oermann EK, Uhm S, Kim JS, Chen LN, Li X, Berry DL, Kallakury BV, Chauhan SC, Collins SP, Suy S, Kumar D. Srivastava A, et al. PLoS One. 2013 Oct 22;8(10):e76994. doi: 10.1371/journal.pone.0076994. eCollection 2013. PLoS One. 2013. PMID: 24167554 Free PMC article. Clinical Trial. - MiR-650 represses high-risk non-metastatic colorectal cancer progression via inhibition of AKT2/GSK3β/E-cadherin pathway.
Zhou C, Cui F, Li J, Wang D, Wei Y, Wu Y, Wang J, Zhu H, Wang S. Zhou C, et al. Oncotarget. 2017 Jul 25;8(30):49534-49547. doi: 10.18632/oncotarget.17743. Oncotarget. 2017. PMID: 28548936 Free PMC article. - The pancreatic tumor microenvironment drives changes in miRNA expression that promote cytokine production and inhibit migration by the tumor associated stroma.
Han S, Gonzalo DH, Feely M, Delitto D, Behrns KE, Beveridge M, Zhang D, Thomas R, Trevino JG, Schmittgen TD, Hughes SJ. Han S, et al. Oncotarget. 2016 Jul 20;8(33):54054-54067. doi: 10.18632/oncotarget.10722. eCollection 2017 Aug 15. Oncotarget. 2016. PMID: 28903323 Free PMC article.
References
- Chandrasekharappa SC, Guru SC, Manickam P, Olufemi SE, Collins FS, Emmert-Buck MR, Debelenko LV, Zhuang Z, Lubensky IA, Liotta LA, Crabtree JS, Wang Y, Roe BA, Weisemann J, Boguski MS, Agarwal SK, Kester MB, Kim YS, Heppner C, Dong Q, Spiegel AM, Burns AL, Marx SJ. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical