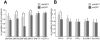KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading - PubMed (original) (raw)
KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading
Anna C Groner et al. PLoS Genet. 2010.
Abstract
Krüppel-associated box domain-zinc finger proteins (KRAB-ZFPs) are tetrapod-specific transcriptional repressors encoded in the hundreds by the human genome. In order to explore their as yet ill-defined impact on gene expression, we developed an ectopic repressor assay, allowing the study of KRAB-mediated transcriptional regulation at hundreds of different transcriptional units. By targeting a drug-controllable KRAB-containing repressor to gene-trapping lentiviral vectors, we demonstrate that KRAB and its corepressor KAP1 can silence promoters located several tens of kilobases (kb) away from their DNA binding sites, with an efficiency which is generally higher for promoters located within 15 kb or less. Silenced promoters exhibit a loss of histone H3-acetylation, an increase in H3 lysine 9 trimethylation (H3K9me3), and a drop in RNA Pol II recruitment, consistent with a block of transcriptional initiation following the establishment of silencing marks. Furthermore, we reveal that KRAB-mediated repression is established by the long-range spreading of H3K9me3 and heterochromatin protein 1 beta (HP1beta) between the repressor binding site and the promoter. We confirm the biological relevance of this phenomenon by documenting KAP1-dependent transcriptional repression at an endogenous KRAB-ZFP gene cluster, where KAP1 binds to the 3' end of genes and mediates propagation of H3K9me3 and HP1beta towards their 5' end. Together, our data support a model in which KRAB/KAP1 recruitment induces long-range repression through the spread of heterochromatin. This finding not only suggests auto-regulatory mechanisms in the control of KRAB-ZFP gene clusters, but also provides important cues for interpreting future genome-wide DNA binding data of KRAB-ZFPs and KAP1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
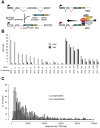
Figure 1. KRAB–mediated silencing can act over several tens of kilobases.
(A) Mechanism of how endogenous genes are targeted by tTRKRAB using the lentiviral vector-based “Trapping/Silencing” (TrapSil) system: _TetO_-containing gene traps carrying the promoterless puroR-GFP gene only express this reporter if after proviral integration they “trap” an actively transcribing gene. The TetO sites further allow binding of the ectopic repressor tTRKRAB to the gene traps after dox removal, while the trap reporter serves as a direct read-out for the effects of tTRKRAB–mediated “silencing”. (B) tTRKRAB–expressing HeLa cells were transduced with LV TrapSil vectors and 23 clones expressing the trap reporter were isolated. Mean fluorescence intensity (MFI) GFP values of these individual clones, cultured with and without dox, were determined and the ratios of these values were used to calculate the silencing efficiency (% silencing = 1- ((MFI GFP –dox)/(MFI GFP +dox))) depicted below the x-axis for each clone. (C) GFP-mediated cell sorting was used to isolate populations of HeLa cells exhibiting either a “repressible” (>90% silencing) or an “irrepressible” (<10% silencing) phenotype. 484 repressible and 699 irrepressible clonal integrations were mapped and the distance between them and their trapped promoter was plotted on a cumulative histogram.
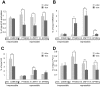
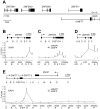
Figure 2. Silenced promoters exhibit increased H3K9me3 and decreased H3-acetylation and RNA Pol II recruitment.
ChIP–based measurement of (A) H3-acetylation, (B) H3K9me3, (C) total histone H3, and (D) hypophosphorylated RNA Pol II at the irrepressible promoter of casein kinase 1d (CSNK1D) and at the repressible promoters of prostaglandin E synthase 3 (PTGES3), zinc finger protein 77 (ZNF77) and ephrin receptor B4 (EPHB4). The relative enrichment values (% of input) of H3-acetylation, total H3, and RNA Pol II were normalized to the relative enrichment at the GAPDH promoter. All values are expressed as means +SEM of triplicate experiments. The statistical test conducted was the student's t-test comparing +dox to –dox samples. If unequal variances were observed, the values were log10 transformed. The criterion for significance for all analyses was p<0.05. *: p<0.05, **, p<0.01, ***, p<0.005.
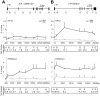
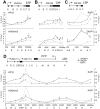
Figure 3. HP1β and H3K9me3 spread from the KRAB–binding site to the promoter.
ChIP analyses quantifying the relative enrichment (% of input) of both H3K9me3 and HP1β were performed for the irrepressible clone XVI and the repressible clone I in the presence and absence of dox. The interrogated sequence spanned from the proviral tTRKRAB binding sites (light grey circles) to the indicated trapped promoters. (A) Enrichments of both modifications at the trapped casein kinase 1d (CSNK1D) locus. (B) Enrichments of both modifications at the prostaglandin E synthase 3 (PTGES3) locus. qPCR amplicons are depicted as letters and are not drawn to scale. All values are expressed as means +SEM of triplicate experiments. Fold changes were calculated as ratios of -dox/+dox enrichments, with the ratios of the respective positive controls set as 1. The controls consisted of p53BP2 for H3K9me3 and of ZNF556 for HP1β (Table S4).
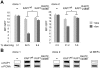
Figure 4. HP1 facilitates KRAB/KAP1–mediated long-range repression.
(A) Two LV TrapSil-selected, tTRKRAB–expressing KAP1−/− MEF clones were complemented by transduction with a lentiviral vector expressing either wild type KAP1 (KAP1wt) or HP1 binding-defective KAP1 (KAP1R487E, V488E), before FACS analysis in the presence or absence of dox. The mean fluorescence intensity values (MFI) of GFP and the silencing efficiency values are depicted as means of six independent measurements. The statistical analysis was performed using two-way ANOVA and, since the interactions were linked, the simple main effects were analyzed. The criterion for significance for all analyses was p<0.05. *: p<0.05. (B) Western blot analysis monitoring KAP1 protein levels, using PCNA as a loading control. The far right panel shows endogenous KAP1 expression in a MEF wild-type cell line.
Figure 5. KAP1 mediates transcriptional repression of KRAB–ZFPs.
mRNA expression levels upon stable KAP1 (shKAP1) and GFP (shGFP) knockdown were assessed through qPCR in HeLa cells. The relative quantities were measured for (A) cluster-specific KRAB–ZFP transcripts and for (B) different heterochromatin protein expression levels. β2-microglobulin was used as a normalization gene, whereas the expression of the eukaryotic elongation factor 1α (EEF1α) served as a negative control. qPCR values are expressed as means +SEM of six experiments. The statistical test conducted was a t-test with Welch's correction, which assumes unequal variances: *: p<0.05.
Figure 6. KAP1 is enriched at the 3′end of KRAB–ZFPs.
(A) Depiction of the KRAB–ZFP cluster under study, located on human chromosome 19. (B–E) Direct KAP1 binding to this cluster was assessed by ChIP for the (B) ZNF554 gene, (C) ZNF555 gene, (D) ZNF556 gene, and (E) the region containing ZNF77 and the overlapping ZNF57 gene. Values are the result of duplicate experiments and are depicted with +SEM error bars.
Figure 7. KAP1 mediates the spread of HP1β and H3K9me3 at KRAB–ZFP clusters.
ChIP-mediated measurement of HP1β and H3K9me3 in control (shGFP) and KAP1 knockdown (shKAP1) HeLa cells at the (A) ZNF554 gene, (B) ZNF555 gene, (C) ZNF556 gene, and at the (D) ZNF77/ZNF57 genes. Dotted lines depict the location of KAP1 peaks determined (see Figure 6B–6E). All values are expressed as means +SEM of duplicate experiments. We ensured that comparable amounts of HP1β and H3K9me3 ChIP material were present in our shKAP1 and shGFP cell line by comparing relative enrichment levels at control loci (Figure S8).
Similar articles
- A gene-rich, transcriptionally active environment and the pre-deposition of repressive marks are predictive of susceptibility to KRAB/KAP1-mediated silencing.
Meylan S, Groner AC, Ambrosini G, Malani N, Quenneville S, Zangger N, Kapopoulou A, Kauzlaric A, Rougemont J, Ciuffi A, Bushman FD, Bucher P, Trono D. Meylan S, et al. BMC Genomics. 2011 Jul 26;12:378. doi: 10.1186/1471-2164-12-378. BMC Genomics. 2011. PMID: 21791101 Free PMC article. - SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins.
Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. Schultz DC, et al. Genes Dev. 2002 Apr 15;16(8):919-32. doi: 10.1101/gad.973302. Genes Dev. 2002. PMID: 11959841 Free PMC article. - Evolution of KRAB-containing zinc finger proteins and their roles in species evolution.
Wang J, Wang J, Tian CY. Wang J, et al. Yi Chuan. 2016 Nov 20;38(11):971-978. doi: 10.16288/j.yczz.16-056. Yi Chuan. 2016. PMID: 27867147 Review. - KRAB zinc finger proteins.
Ecco G, Imbeault M, Trono D. Ecco G, et al. Development. 2017 Aug 1;144(15):2719-2729. doi: 10.1242/dev.132605. Development. 2017. PMID: 28765213 Free PMC article. Review.
Cited by
- Human Endogenous Retrovirus as Therapeutic Targets in Neurologic Disease.
Giménez-Orenga K, Oltra E. Giménez-Orenga K, et al. Pharmaceuticals (Basel). 2021 May 24;14(6):495. doi: 10.3390/ph14060495. Pharmaceuticals (Basel). 2021. PMID: 34073730 Free PMC article. Review. - Visualizing complex feature interactions and feature sharing in genomic deep neural networks.
Liu G, Zeng H, Gifford DK. Liu G, et al. BMC Bioinformatics. 2019 Jul 19;20(1):401. doi: 10.1186/s12859-019-2957-4. BMC Bioinformatics. 2019. PMID: 31324140 Free PMC article. - Genome-wide functional perturbation of human microsatellite repeats using engineered zinc finger transcription factors.
Tak YE, Boulay G, Lee L, Iyer S, Perry NT, Schultz HT, Garcia SP, Broye L, Horng JE, Rengarajan S, Naigles B, Volorio A, Sander JD, Gong J, Riggi N, Joung JK, Rivera MN. Tak YE, et al. Cell Genom. 2022 Apr 13;2(4):100119. doi: 10.1016/j.xgen.2022.100119. Cell Genom. 2022. PMID: 35967079 Free PMC article. - Current Approaches to Epigenetic Therapy.
Griazeva ED, Fedoseeva DM, Radion EI, Ershov PV, Meshkov IO, Semyanihina AV, Makarova AS, Makarov VV, Yudin VS, Keskinov AA, Kraevoy SA. Griazeva ED, et al. Epigenomes. 2023 Sep 30;7(4):23. doi: 10.3390/epigenomes7040023. Epigenomes. 2023. PMID: 37873808 Free PMC article. Review. - Gene expression profiling of giant cell tumor of bone reveals downregulation of extracellular matrix components decorin and lumican associated with lung metastasis.
Lieveld M, Bodson E, De Boeck G, Nouman B, Cleton-Jansen AM, Korsching E, Benassi MS, Picci P, Sys G, Poffyn B, Athanasou NA, Hogendoorn PC, Forsyth RG. Lieveld M, et al. Virchows Arch. 2014 Dec;465(6):703-13. doi: 10.1007/s00428-014-1666-7. Epub 2014 Oct 11. Virchows Arch. 2014. PMID: 25304290
References
- Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous