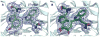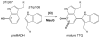In crystallo posttranslational modification within a MauG/pre-methylamine dehydrogenase complex - PubMed (original) (raw)
In crystallo posttranslational modification within a MauG/pre-methylamine dehydrogenase complex
Lyndal M R Jensen et al. Science. 2010.
Abstract
MauG is a diheme enzyme responsible for the posttranslational modification of two tryptophan residues to form the tryptophan tryptophylquinone (TTQ) cofactor of methylamine dehydrogenase (MADH). MauG converts preMADH, containing monohydroxylated betaTrp57, to fully functional MADH by catalyzing the insertion of a second oxygen atom into the indole ring and covalently linking betaTrp57 to betaTrp108. We have solved the x-ray crystal structure of MauG complexed with preMADH to 2.1 angstroms. The c-type heme irons and the nascent TTQ site are separated by long distances over which electron transfer must occur to achieve catalysis. In addition, one of the hemes has an atypical His-Tyr axial ligation. The crystalline protein complex is catalytically competent; upon addition of hydrogen peroxide, MauG-dependent TTQ synthesis occurs.
Figures
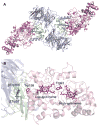
Figure 1
(A) Overall ribbon representation of the MauG-preMADH complex. (B) Spatial layout of potential redox groups. Color scheme is MauG (pink); preMADH α (blue) and β (green). The hemes, Trp93, Trp199 of MauG and βTrp108 and mono-hydroxylated βTrp57 of preMADH are drawn explicitly in a stick representation. Figure produced using PyMOL (
).
Figure 2
Site of TTQ formation in MADH. (A) 2Fo-Fc electron density for the MauG-preMADH complex (resolution 2.1 Å). (B) The first 2Fo-Fc electron density calculated with MauG-preMADH + H2O2 structure factors (resolution 2.1 Å) and MauG-preMADH model phases with the preMADH βTrp57 and βTrp108 side-chains omitted. Electron densities were contoured at 1 σ. Carbon coloring: preMADH, light green; preMADH + H2O2, dark green. Figure produced using PyMOL (
).
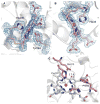
Figure 3
MauG hemes. (A) 6-coordinate low-spin heme (CHE600), (B) 5-coordinate high-spin heme (CHE500), and (C) residues that line the distal pocket. 2Fo-Fc electron density contoured at 1 σ. Figure produced using PyMOL (
).
Scheme 1
Overall reaction catalyzed by MauG
Comment in
- Biochemistry. Remote enzyme microsurgery.
Bollinger JM Jr, Matthews ML. Bollinger JM Jr, et al. Science. 2010 Mar 12;327(5971):1337-8. doi: 10.1126/science.1187421. Science. 2010. PMID: 20223975 No abstract available.
Similar articles
- Posttranslational biosynthesis of the protein-derived cofactor tryptophan tryptophylquinone.
Davidson VL, Wilmot CM. Davidson VL, et al. Annu Rev Biochem. 2013;82:531-50. doi: 10.1146/annurev-biochem-051110-133601. Annu Rev Biochem. 2013. PMID: 23746262 Free PMC article. Review. - Crystal structures of CO and NO adducts of MauG in complex with pre-methylamine dehydrogenase: implications for the mechanism of dioxygen activation.
Yukl ET, Goblirsch BR, Davidson VL, Wilmot CM. Yukl ET, et al. Biochemistry. 2011 Apr 12;50(14):2931-8. doi: 10.1021/bi200023n. Epub 2011 Mar 16. Biochemistry. 2011. PMID: 21355604 Free PMC article. - Tryptophan tryptophylquinone biosynthesis: a radical approach to posttranslational modification.
Davidson VL, Liu A. Davidson VL, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1299-305. doi: 10.1016/j.bbapap.2012.01.008. Epub 2012 Jan 28. Biochim Biophys Acta. 2012. PMID: 22314272 Free PMC article. Review. - Mutagenesis of tryptophan199 suggests that hopping is required for MauG-dependent tryptophan tryptophylquinone biosynthesis.
Tarboush NA, Jensen LM, Yukl ET, Geng J, Liu A, Wilmot CM, Davidson VL. Tarboush NA, et al. Proc Natl Acad Sci U S A. 2011 Oct 11;108(41):16956-61. doi: 10.1073/pnas.1109423108. Epub 2011 Oct 3. Proc Natl Acad Sci U S A. 2011. PMID: 21969534 Free PMC article. - Proline 107 is a major determinant in maintaining the structure of the distal pocket and reactivity of the high-spin heme of MauG.
Feng M, Jensen LM, Yukl ET, Wei X, Liu A, Wilmot CM, Davidson VL. Feng M, et al. Biochemistry. 2012 Feb 28;51(8):1598-606. doi: 10.1021/bi201882e. Epub 2012 Feb 10. Biochemistry. 2012. PMID: 22299652 Free PMC article.
Cited by
- MauG: a di-heme enzyme required for methylamine dehydrogenase maturation.
Wilmot CM, Yukl ET. Wilmot CM, et al. Dalton Trans. 2013 Mar 7;42(9):3127-35. doi: 10.1039/c2dt32059b. Epub 2012 Oct 22. Dalton Trans. 2013. PMID: 23086017 Free PMC article. Review. - Posttranslational biosynthesis of the protein-derived cofactor tryptophan tryptophylquinone.
Davidson VL, Wilmot CM. Davidson VL, et al. Annu Rev Biochem. 2013;82:531-50. doi: 10.1146/annurev-biochem-051110-133601. Annu Rev Biochem. 2013. PMID: 23746262 Free PMC article. Review. - A Stable Ferryl Porphyrin at the Active Site of Y463M BthA.
Rizzolo K, Weitz AC, Cohen SE, Drennan CL, Hendrich MP, Elliott SJ. Rizzolo K, et al. J Am Chem Soc. 2020 Jul 15;142(28):11978-11982. doi: 10.1021/jacs.0c04023. Epub 2020 Jul 1. J Am Chem Soc. 2020. PMID: 32564595 Free PMC article. - Effects of the loss of the axial tyrosine ligand of the low-spin heme of MauG on its physical properties and reactivity.
Abu Tarboush N, Shin S, Geng J, Liu A, Davidson VL. Abu Tarboush N, et al. FEBS Lett. 2012 Dec 14;586(24):4339-43. doi: 10.1016/j.febslet.2012.10.044. Epub 2012 Nov 2. FEBS Lett. 2012. PMID: 23127557 Free PMC article. - A T67A mutation in the proximal pocket of the high-spin heme of MauG stabilizes formation of a mixed-valent FeII/FeIII state and enhances charge resonance stabilization of the bis-FeIV state.
Shin S, Feng M, Li C, Williamson HR, Choi M, Wilmot CM, Davidson VL. Shin S, et al. Biochim Biophys Acta. 2015 Aug;1847(8):709-16. doi: 10.1016/j.bbabio.2015.04.008. Epub 2015 Apr 17. Biochim Biophys Acta. 2015. PMID: 25896561 Free PMC article.
References
- Davidson VL. Biochemistry. 2007 May 8;46:5283. - PubMed
- Wang Y, et al. Biochemistry. 2003 Jun 24;42:7318. - PubMed
- McIntire WS, Wemmer DE, Chistoserdov A, Lidstrom ME. Science. 1991 May 10;252:817. - PubMed
- Davidson VL. Adv Protein Chem. 2001;58:95. - PubMed
- Chen LY, et al. J Mol Biol. 1998 Feb 13;276:131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- R37 GM041574/GM/NIGMS NIH HHS/United States
- R01 GM066569-08/GM/NIGMS NIH HHS/United States
- GM66569/GM/NIGMS NIH HHS/United States
- R01 GM041574/GM/NIGMS NIH HHS/United States
- R01 GM066569/GM/NIGMS NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
- GM41574/GM/NIGMS NIH HHS/United States
- R01 GM041574-20/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources