FastTree 2--approximately maximum-likelihood trees for large alignments - PubMed (original) (raw)
FastTree 2--approximately maximum-likelihood trees for large alignments
Morgan N Price et al. PLoS One. 2010.
Abstract
Background: We recently described FastTree, a tool for inferring phylogenies for alignments with up to hundreds of thousands of sequences. Here, we describe improvements to FastTree that improve its accuracy without sacrificing scalability.
Methodology/principal findings: Where FastTree 1 used nearest-neighbor interchanges (NNIs) and the minimum-evolution criterion to improve the tree, FastTree 2 adds minimum-evolution subtree-pruning-regrafting (SPRs) and maximum-likelihood NNIs. FastTree 2 uses heuristics to restrict the search for better trees and estimates a rate of evolution for each site (the "CAT" approximation). Nevertheless, for both simulated and genuine alignments, FastTree 2 is slightly more accurate than a standard implementation of maximum-likelihood NNIs (PhyML 3 with default settings). Although FastTree 2 is not quite as accurate as methods that use maximum-likelihood SPRs, most of the splits that disagree are poorly supported, and for large alignments, FastTree 2 is 100-1,000 times faster. FastTree 2 inferred a topology and likelihood-based local support values for 237,882 distinct 16S ribosomal RNAs on a desktop computer in 22 hours and 5.8 gigabytes of memory.
Conclusions/significance: FastTree 2 allows the inference of maximum-likelihood phylogenies for huge alignments. FastTree 2 is freely available at http://www.microbesonline.org/fasttree.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
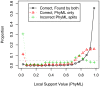
Figure 1. Local support values for splits found by PhyML with SPR moves and/or FastTree.
We examined local support values for the splits inferred by PhyML 3.0 with  + SPRs on simulated alignments with 250 protein sequences. We classified PhyML's splits as correct and found by both PhyML and FastTree, correct but missed by FastTree, or incorrect. We show the distribution of support values for each class. The right-most bin includes the strongly supported splits (0.95 to 1.0), and the gray dashed line shows the uniform distribution. The support values are PhyML's minimum of the approximate likelihood ratio test and SH-like , local supports.
+ SPRs on simulated alignments with 250 protein sequences. We classified PhyML's splits as correct and found by both PhyML and FastTree, correct but missed by FastTree, or incorrect. We show the distribution of support values for each class. The right-most bin includes the strongly supported splits (0.95 to 1.0), and the gray dashed line shows the uniform distribution. The support values are PhyML's minimum of the approximate likelihood ratio test and SH-like , local supports.
Figure 2. Likelihoods over time for genuine alignments.
Each line shows the time it takes a different tool to reach a given likelihood. For the COG alignments, all times and likelihoods are averages over the seven alignments. For FastTree, we show the time and the improvement in likelihood for the minimum-evolution topology and the final (approximately-ML) topology. For RAxML, we show the maximum parsimony starting topology, the first two rounds of SPR moves, and the final topology (note the interrupted  axis). For RAxML with FastTree's (minimum-evolution) starting tree, we show the starting topology and RAxML's first two rounds of SPR moves.
axis). For RAxML with FastTree's (minimum-evolution) starting tree, we show the starting topology and RAxML's first two rounds of SPR moves.
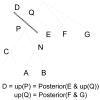
Figure 3. Traversing a tree with up-posteriors.
FastTree optimizes the tree near node N by analyzing the posterior distributions for subtrees A, B, and C, as well as the “up-posterior” D.
Similar articles
- FastTree: computing large minimum evolution trees with profiles instead of a distance matrix.
Price MN, Dehal PS, Arkin AP. Price MN, et al. Mol Biol Evol. 2009 Jul;26(7):1641-50. doi: 10.1093/molbev/msp077. Epub 2009 Apr 17. Mol Biol Evol. 2009. PMID: 19377059 Free PMC article. - RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation.
Liu K, Linder CR, Warnow T. Liu K, et al. PLoS One. 2011;6(11):e27731. doi: 10.1371/journal.pone.0027731. Epub 2011 Nov 21. PLoS One. 2011. PMID: 22132132 Free PMC article. - morePhyML: improving the phylogenetic tree space exploration with PhyML 3.
Criscuolo A. Criscuolo A. Mol Phylogenet Evol. 2011 Dec;61(3):944-8. doi: 10.1016/j.ympev.2011.08.029. Epub 2011 Sep 8. Mol Phylogenet Evol. 2011. PMID: 21925283 - Very Fast Tree: speeding up the estimation of phylogenies for large alignments through parallelization and vectorization strategies.
Piñeiro C, Abuín JM, Pichel JC. Piñeiro C, et al. Bioinformatics. 2020 Nov 1;36(17):4658-4659. doi: 10.1093/bioinformatics/btaa582. Bioinformatics. 2020. PMID: 32573652 - New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. Guindon S, et al. Syst Biol. 2010 May;59(3):307-21. doi: 10.1093/sysbio/syq010. Epub 2010 Mar 29. Syst Biol. 2010. PMID: 20525638
Cited by
- Microbial diversity in the deep-subsurface hydrothermal aquifer feeding the giant gypsum crystal-bearing Naica Mine, Mexico.
Ragon M, Van Driessche AE, García-Ruíz JM, Moreira D, López-García P. Ragon M, et al. Front Microbiol. 2013 Mar 6;4:37. doi: 10.3389/fmicb.2013.00037. eCollection 2013. Front Microbiol. 2013. PMID: 23508882 Free PMC article. - A cross-taxon analysis of insect-associated bacterial diversity.
Jones RT, Sanchez LG, Fierer N. Jones RT, et al. PLoS One. 2013 Apr 16;8(4):e61218. doi: 10.1371/journal.pone.0061218. Print 2013. PLoS One. 2013. PMID: 23613815 Free PMC article. - Whole genome sequencing of Streptomyces actuosus ISP-5337, Streptomyces sioyaensis B-5408, and Actinospica acidiphila B-2296 reveals secondary metabolomes with antibiotic potential.
Majer HM, Ehrlich RL, Ahmed A, Earl JP, Ehrlich GD, Beld J. Majer HM, et al. Biotechnol Rep (Amst). 2021 Feb 9;29:e00596. doi: 10.1016/j.btre.2021.e00596. eCollection 2021 Mar. Biotechnol Rep (Amst). 2021. PMID: 33643857 Free PMC article. - Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae).
Andersson MN, Grosse-Wilde E, Keeling CI, Bengtsson JM, Yuen MM, Li M, Hillbur Y, Bohlmann J, Hansson BS, Schlyter F. Andersson MN, et al. BMC Genomics. 2013 Mar 21;14:198. doi: 10.1186/1471-2164-14-198. BMC Genomics. 2013. PMID: 23517120 Free PMC article. - Mobile Element Integration Reveals a Chromosome Dimer Resolution System in Legionellales.
Nicholson B, Deecker SR, Ensminger AW. Nicholson B, et al. mBio. 2022 Dec 20;13(6):e0217122. doi: 10.1128/mbio.02171-22. Epub 2022 Oct 31. mBio. 2022. PMID: 36314797 Free PMC article.
References
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. - PubMed
- Studier JA, Keppler KJ. A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol. 1988;5:729–31. - PubMed
- Felsenstein J. Evolutionary trees from dna sequences: A maximum likelihood approach. J Mol Evol. 1981;17:368–376. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous