Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening - PubMed (original) (raw)
Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening
Kianoush Sadre-Bazzaz et al. Mol Cell. 2010.
Abstract
The proteasome is an abundant protease that is critically important for numerous cellular pathways. Proteasomes are activated in vitro by three known classes of proteins/complexes, including Blm10/PA200. Here, we report a 3.4 A resolution crystal structure of a proteasome-Blm10 complex, which reveals that Blm10 surrounds the proteasome entry pore in the 1.2 MDa complex to form a largely closed dome that is expected to restrict access of potential substrates. This architecture and the observation that Blm10 induces a disordered proteasome gate structure challenge the assumption that Blm10 functions as an activator of proteolysis in vivo. The Blm10 C terminus binds in the same manner as seen for 11S activators and inferred for 19S/PAN activators and indicates a unified model for gate opening. We also demonstrate that Blm10 acts to maintain mitochondrial function. Consistent with the structural data, the C-terminal residues of Blm10 are needed for this activity.
(c) 2010 Elsevier Inc. All rights reserved.
Figures
Figure 1. Structure of the Blm10:proteasome complex
(A) Cartoon of the proteasome-Blm10 complex, side view. Proteasome white (α subunits) and gray (β subunits), Blm10 rainbow from N-terminus (blue) to C-terminus (red). (B) Cutaway view with the molecular surface. (C) Electron density for Blm10. All maps shown in this paper were phased on the proteasome molecular replacement model for which segments approaching Blm10 had been removed. The phases were refined by solvent flattening, histogram shifting, and four-fold non-crystallographic symmetry averaging. (D) Top view, space-filling representation. The opening visible in the center of this view measures only ~6Å between atom centers. The largest opening, which is not visible in this orientation, is indicated with an arrow. (E) Same as panel D, but only showing Blm10 residues that have at least one atom within 6 Å of the proteasome. (F) Close-up view of the largest opening through the Blm10 dome. Orientation as indicated by the arrow in panel D. Asterisks denote the last ordered residues adjacent to disordered segments that have been omitted from the model.
Figure 2. Proteasome conformational changes induced by Blm10
(A) Top view of the proteasome pore region with electron density for the Blm10 complex. The absence of density for the N-terminal residues of proteasome α2, α3, and α4 indicates that they are disordered in the Blm10 complex (white), whereas they are ordered in the closed, unliganded conformation (colors) and in the fully open complex with PA26 (not shown in this panel). (B) Open conformation seen in complexes with PA26 (yellow) and Blm10 (white). The stabilizing cluster residues (Tyr8, Asp9, Pro17, Tyr26; (Forster et al., 2003)) are labeled for the α6/α7 cluster, which is ordered in the unliganded proteasome (Groll et al., 1997) and in both the PA26 and Blm10 complexes shown here. Tyr8 and Asp9 residues are not ordered for α2, α3, or α4 in the Blm10 complex. Residues indicated with an asterisk are ordered in the Blm10 complex but are displaced from the open conformation seen with PA26. A version of this panel that also includes the closed conformation is shown in Figure S2. (C) Contacts that stabilize α5Asp9 away from the open conformation. (D) Contacts that stabilize α7Tyr8 away from the open conformation.
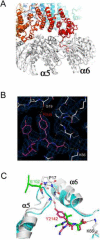
Figure 3. Interactions of the Blm10 C-terminal residues
(A) Side view with Blm10 C-terminus labeled “C”. (B) The electron density map is well defined for the Blm10 penultimate tyrosine (Tyr2142) and surrounding residues. (C) The last three residues of PA26 (green) and Blm10 (red) are shown after overlap of the two complexes on surrounding proteasome residues. Unliganded proteasome (Groll et al., 1997), cyan. Blm10 Tyr2142 stabilizes the open position of α5 by hydrogen bonding with Gly19 O. PA26 stabilizes the same transition by hydrogen bonding interactions of its activation loop residue Glu102.
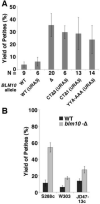
Figure 4. The C-terminal residues of Blm10 are important for its function
(A) Isogenic strains were constructed in the A364a genetic background with the genotypes indicated (Table S1). Multiple independent colonies of each strain growing on glycerol medium to select for retention of mitochondrial function were used to inoculate rich medium containing glucose. Saturated cultures were diluted and plated on rich glucose medium, then mitochondrial function in clones was assessed using the tetrazolium staining method (Ogur et al., 1957). Results from multiple strains with the same genotype were combined (Table S1, total number indicated as N), with the average percentage yield of petite colonies plotted here. Error bars indicate the standard deviation of the measurements. (B) As in panel A, except isogenic pairs from three other commonly used genetic backgrounds containing or lacking Blm10 were assayed by picking 120 colonies without regard to size, then replica plating to media containing glycerol or glucose to determine the number of petite colonies (Table S1). See also Figures S3.
Similar articles
- Proteasome activators.
Stadtmueller BM, Hill CP. Stadtmueller BM, et al. Mol Cell. 2011 Jan 7;41(1):8-19. doi: 10.1016/j.molcel.2010.12.020. Mol Cell. 2011. PMID: 21211719 Free PMC article. Review. - Structure of the Blm10-20 S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes.
Iwanczyk J, Sadre-Bazzaz K, Ferrell K, Kondrashkina E, Formosa T, Hill CP, Ortega J. Iwanczyk J, et al. J Mol Biol. 2006 Oct 27;363(3):648-59. doi: 10.1016/j.jmb.2006.08.010. Epub 2006 Aug 9. J Mol Biol. 2006. PMID: 16952374 Free PMC article. - The Proteasome Activators Blm10/PA200 Enhance the Proteasomal Degradation of N-Terminal Huntingtin.
Aladdin A, Yao Y, Yang C, Kahlert G, Ghani M, Király N, Boratkó A, Uray K, Dittmar G, Tar K. Aladdin A, et al. Biomolecules. 2020 Nov 20;10(11):1581. doi: 10.3390/biom10111581. Biomolecules. 2020. PMID: 33233776 Free PMC article. - Blm10 binds to pre-activated proteasome core particles with open gate conformation.
Lehmann A, Jechow K, Enenkel C. Lehmann A, et al. EMBO Rep. 2008 Dec;9(12):1237-43. doi: 10.1038/embor.2008.190. Epub 2008 Oct 17. EMBO Rep. 2008. PMID: 18927584 Free PMC article. - Proteasome activator 200: the heat is on..
Savulescu AF, Glickman MH. Savulescu AF, et al. Mol Cell Proteomics. 2011 May;10(5):R110.006890. doi: 10.1074/mcp.R110.006890. Epub 2011 Mar 9. Mol Cell Proteomics. 2011. PMID: 21389348 Free PMC article. Review.
Cited by
- Proteasome allostery as a population shift between interchanging conformers.
Ruschak AM, Kay LE. Ruschak AM, et al. Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):E3454-62. doi: 10.1073/pnas.1213640109. Epub 2012 Nov 12. Proc Natl Acad Sci U S A. 2012. PMID: 23150576 Free PMC article. - Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome.
Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rosenzweig R, et al. J Biol Chem. 2012 Apr 27;287(18):14659-71. doi: 10.1074/jbc.M111.316323. Epub 2012 Feb 8. J Biol Chem. 2012. PMID: 22318722 Free PMC article. - Chaperone-mediated assembly of the proteasome core particle - recent developments and structural insights.
Schnell HM, Walsh RM, Rawson S, Hanna J. Schnell HM, et al. J Cell Sci. 2022 Apr 15;135(8):jcs259622. doi: 10.1242/jcs.259622. Epub 2022 Apr 22. J Cell Sci. 2022. PMID: 35451017 Free PMC article. Review. - Structure characterization of the 26S proteasome.
Kim HM, Yu Y, Cheng Y. Kim HM, et al. Biochim Biophys Acta. 2011 Feb;1809(2):67-79. doi: 10.1016/j.bbagrm.2010.08.008. Epub 2010 Aug 26. Biochim Biophys Acta. 2011. PMID: 20800708 Free PMC article. Review. - Proteasome activators.
Stadtmueller BM, Hill CP. Stadtmueller BM, et al. Mol Cell. 2011 Jan 7;41(1):8-19. doi: 10.1016/j.molcel.2010.12.020. Mol Cell. 2011. PMID: 21211719 Free PMC article. Review.
References
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. - PubMed
- Blickwedehl J, McEvoy S, Wong I, Kousis P, Clements J, Elliott R, Cresswell P, Liang P, Bangia N. Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiat Res. 2007;167:663–674. - PubMed
- Doherty K, Pramanik A, Pride L, Lukose J, Moore CW. Expression of the expanded YFL007w ORF and assignment of the gene name BLM10. Yeast. 2004;21:1021–1023. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 EB009998/EB/NIBIB NIH HHS/United States
- R01 GM059135/GM/NIGMS NIH HHS/United States
- R01 GM059135-09A1/GM/NIGMS NIH HHS/United States
- R01 GM059135-11/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases