Diversity of anaerobic microbes in spacecraft assembly clean rooms - PubMed (original) (raw)
Diversity of anaerobic microbes in spacecraft assembly clean rooms
Alexander Probst et al. Appl Environ Microbiol. 2010 May.
Abstract
Although the cultivable and noncultivable microbial diversity of spacecraft assembly clean rooms has been previously documented using conventional and state-of-the-art molecular techniques, the occurrence of obligate anaerobes within these clean rooms is still uncertain. Therefore, anaerobic bacterial communities of three clean-room facilities were analyzed during assembly of the Mars Science Laboratory rover. Anaerobic bacteria were cultured on several media, and DNA was extracted from suitable anaerobic enrichments and examined with conventional 16S rRNA gene clone library, as well as high-density phylogenetic 16S rRNA gene microarray (PhyloChip) technologies. The culture-dependent analyses predominantly showed the presence of clostridial and propionibacterial strains. The 16S rRNA gene sequences retrieved from clone libraries revealed distinct microbial populations associated with each clean-room facility, clustered exclusively within gram-positive organisms. PhyloChip analysis detected a greater microbial diversity, spanning many phyla of bacteria, and provided a deeper insight into the microbial community structure of the clean-room facilities. This study presents an integrated approach for assessing the anaerobic microbial population within clean-room facilities, using both molecular and cultivation-based analyses. The results reveal that highly diverse anaerobic bacterial populations persist in the clean rooms even after the imposition of rigorous maintenance programs and will pose a challenge to planetary protection implementation activities.
Figures
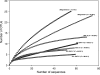
FIG. 1.
Rarefaction analysis of anaerobic bacterial diversity from several spacecraft assembly facilities. The coverage values (C) for each clone library are given in parentheses. Rarefaction curves of standard and gradient PCR samples are depicted with asterisks and in italics, respectively.
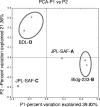
FIG. 2.
Principal coordinate analyses showing the relationships between samples.
FIG. 3.
Overview of phyla retrieved from the DNA microarray analysis. Numbers in parentheses on the x axis after each sampling event are the number of OTU counts retrieved. Families having a minimum of 10 OTU counts are given in parentheses. Underlined families were also detected in conventional clone libraries of related samplings. The “others” category denotes the phyla having fewer than 10 OTU counts as well as unclassified members.
Similar articles
- Cultivation of anaerobic and facultatively anaerobic bacteria from spacecraft-associated clean rooms.
Stieglmeier M, Wirth R, Kminek G, Moissl-Eichinger C. Stieglmeier M, et al. Appl Environ Microbiol. 2009 Jun;75(11):3484-91. doi: 10.1128/AEM.02565-08. Epub 2009 Apr 10. Appl Environ Microbiol. 2009. PMID: 19363082 Free PMC article. - The first collection of spacecraft-associated microorganisms: a public source for extremotolerant microorganisms from spacecraft assembly clean rooms.
Moissl-Eichinger C, Rettberg P, Pukall R. Moissl-Eichinger C, et al. Astrobiology. 2012 Nov;12(11):1024-34. doi: 10.1089/ast.2012.0906. Astrobiology. 2012. PMID: 23121015 - Comprehensive census of bacteria in clean rooms by using DNA microarray and cloning methods.
La Duc MT, Osman S, Vaishampayan P, Piceno Y, Andersen G, Spry JA, Venkateswaran K. La Duc MT, et al. Appl Environ Microbiol. 2009 Oct;75(20):6559-67. doi: 10.1128/AEM.01073-09. Epub 2009 Aug 21. Appl Environ Microbiol. 2009. PMID: 19700540 Free PMC article. - Lessons learned from the microbial analysis of the Herschel spacecraft during assembly, integration, and test operations.
Moissl-Eichinger C, Pukall R, Probst AJ, Stieglmeier M, Schwendner P, Mora M, Barczyk S, Bohmeier M, Rettberg P. Moissl-Eichinger C, et al. Astrobiology. 2013 Dec;13(12):1125-39. doi: 10.1089/ast.2013.1024. Epub 2013 Dec 6. Astrobiology. 2013. PMID: 24313230 Review. - Molecular phylogenetic identification of the intestinal anaerobic microbial community in the hindgut of the termite, Reticulitermes speratus, without cultivation.
Kudo T, Ohkuma M, Moriya S, Noda S, Ohtoko K. Kudo T, et al. Extremophiles. 1998 Aug;2(3):155-61. doi: 10.1007/s007920050055. Extremophiles. 1998. PMID: 9783160 Review.
Cited by
- Phenotypic and genomic assessment of the potential threat of human spaceflight-relevant Staphylococcus capitis isolates under stress conditions.
Siems K, Runzheimer K, Rehm A, Schwengers O, Heidler von Heilborn D, Kaser L, Arndt F, Neidhöfer C, Mengel JP, Parcina M, Lipski A, Hain T, Moeller R. Siems K, et al. Front Microbiol. 2022 Nov 3;13:1007143. doi: 10.3389/fmicb.2022.1007143. eCollection 2022. Front Microbiol. 2022. PMID: 36406458 Free PMC article. - Biological Contamination Prevention for Outer Solar System Moons of Astrobiological Interest: What Do We Need to Know?
Rettberg P, Antunes A, Brucato J, Cabezas P, Collins G, Haddaji A, Kminek G, Leuko S, McKenna-Lawlor S, Moissl-Eichinger C, Fellous JL, Olsson-Francis K, Pearce D, Rabbow E, Royle S, Saunders M, Sephton M, Spry A, Walter N, Wimmer Schweingruber R, Treuet JC. Rettberg P, et al. Astrobiology. 2019 Aug;19(8):951-974. doi: 10.1089/ast.2018.1996. Epub 2019 Feb 14. Astrobiology. 2019. PMID: 30762429 Free PMC article. - Performance of Multiple Metagenomics Pipelines in Understanding Microbial Diversity of a Low-Biomass Spacecraft Assembly Facility.
Wood JM, Singh NK, Guan L, Seuylemezian A, Benardini JN, Venkateswaran K. Wood JM, et al. Front Microbiol. 2021 Sep 28;12:685254. doi: 10.3389/fmicb.2021.685254. eCollection 2021. Front Microbiol. 2021. PMID: 34650522 Free PMC article. - Evaluation of procedures for the collection, processing, and analysis of biomolecules from low-biomass surfaces.
Kwan K, Cooper M, La Duc MT, Vaishampayan P, Stam C, Benardini JN, Scalzi G, Moissl-Eichinger C, Venkateswaran K. Kwan K, et al. Appl Environ Microbiol. 2011 May;77(9):2943-53. doi: 10.1128/AEM.02978-10. Epub 2011 Mar 11. Appl Environ Microbiol. 2011. PMID: 21398492 Free PMC article. - A viability-linked metagenomic analysis of cleanroom environments: eukarya, prokaryotes, and viruses.
Weinmaier T, Probst AJ, La Duc MT, Ciobanu D, Cheng JF, Ivanova N, Rattei T, Vaishampayan P. Weinmaier T, et al. Microbiome. 2015 Dec 8;3:62. doi: 10.1186/s40168-015-0129-y. Microbiome. 2015. PMID: 26642878 Free PMC article.
References
- Atlas, R. 1993. Handbook of microbiological media. CRC Press, Boca Raton, FL.
- Brodie, E. L., T. Z. DeSantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. - PMC - PubMed
- COSPAR. 2002. COSPAR planetary protection policy, October 2002, as amended, March 2005. Committee of Space Research 2002, Houston, TX.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous