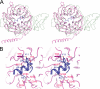Crystal structure of yeast rpn14, a chaperone of the 19 S regulatory particle of the proteasome - PubMed (original) (raw)
Crystal structure of yeast rpn14, a chaperone of the 19 S regulatory particle of the proteasome
Sangwoo Kim et al. J Biol Chem. 2010.
Abstract
The ubiquitin-proteasome pathway is a major proteolytic system in eukaryotic cells and regulates various cellular processes. The 26 S proteasome, the central enzyme of this pathway, consists of a proteolytic core particle and two 19 S regulatory particles (RPs) composed of ATPase (Rpt) and non-ATPase (Rpn) subunits. Growing evidence indicates that proteasome assembly is assisted by a variety of chaperones. In particular, it has been reported recently that Nas2, Nas6, Rpn14, and Hsm3 bind specific Rpt subunits, thereby contributing to the formation of 19 S RP. Rpn14 transiently binds to the C-terminal domain of the Rpt6 subunit (Rpt6-C) during maturation of the ATPase ring of 19 S RP. In this study, we determined the crystal structure of yeast Rpn14 at 2.0 A resolution, which revealed that this chaperone consists of a unique N-terminal domain with unknown function and a C-terminal domain assuming a canonical seven-bladed beta-propeller fold. The Rpt6-binding site on Rpn14 was predicted based on structural comparison with the complex formed between Nas6 and Rpt3-C. The top face of Rpn14 exhibits a highly acidic surface area, whereas the putative interacting surface of Rpt6-C is basic. By inspection of structural data along with genetic and biochemical data, we determined the specific residues of Rpn14 and Rpt6 for complementary charge interactions that are required for 19 S RP assembly.
Figures
FIGURE 1.
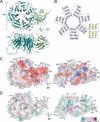
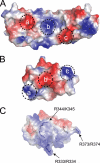
Three-dimensional structure of yeast Rpn14. A, ribbon diagram of Rpn14. The N-terminal domain and the C-terminal β-propeller are colored green and cyan, respectively. The secondary structural elements are labeled. B, topological diagram of the secondary structural elements of Rpn14. The α-helices and β-strands are represented as cylinders and arrows, respectively. C, surface potential representation of Rpn14. Red, blue, and white represent acidic, basic, and neutral, respectively. Mutations are depicted via black boxes around acidic residues. D, molecular surface conservation of Rpn14. Mutation positions of conserved residues are shown by arrows.
FIGURE 2.
Comparison with other WD40 repeat proteins. A, superposition of the β-propellers of Rpn14 (green) and the G protein β-subunit (red). B, close-up view of the interface between the substrate-binding sites of the β-propeller of the G protein β-subunit (red) and the α-helical peptide (blue). Hydrogen bonds are represented by dotted lines.
FIGURE 3.
Comparison of the charge distribution on the surfaces of Nas6, Rpt3-C, Rpn14, and a Rpt6-C homology model based on the Rpt3-C structure. A and B, surface potential representations of Nas6 and Rpt3-C, respectively. The complementary surface patches (a–c) responsible for complex formation are depicted by circles. C, surface potential representation of Rpt6-C. Red, blue, and white represent acidic, basic, and neutral, respectively.
FIGURE 4.
Binding of Rpn14 to the basic residues of the Rpt6 C-terminal region. A, pulldown assay using Rpt6 mutants with Rpn14. His6-tagged Rpn14 was coexpressed with untagged Rpt6-C or its mutants in E. coli and pulled down on nickel-nitrilotriacetic acid resin. The resin-bound proteins were eluted by imidazole-containing buffer, and fractions were analyzed by SDS-PAGE, followed by Coomassie Blue staining. B, genetic interaction between the rpt6R373A/R374A mutation and the NAS6 deletion. Ten serial dilutions of the indicated strains were spotted onto a YPD plate and incubated for 2 days at 25 or 37 °C. C, expression of Nas6, Rpn14, and Rpt6 in mutant cells. Exponentially growing cells in YPD medium were shifted to 37 °C for 4.5 h. Extracts were prepared by the mild alkali method and analyzed by Western blotting with the anti-Nas6, anti-Rpn14, and anti-Rpt6 antibodies. D, native PAGE analysis of the 26 S proteasome assembly in mutant cells. The indicated cells expressing Rpn1-yEGFP1F were cultured as described for C. Extracts were subjected to native PAGE. After electrophoresis, Rpn1-containing complexes or proteasome peptidase activities on the gel were imaged using fluoroimagers. RP2CP and RP1CP indicate singly and doubly capped 26 S proteasomes. Rpn and Rpt subunits are abbreviated as n and t, respectively.
FIGURE 5.
Impact of negatively charged residues on Rpn14 function. A and B, genetic analyses of Rpn14 surface mutants. The Δ_nas6_ Δ_rpn14_ cells harboring a plasmid that expresses RPN14 or mutants under the control of the galactose-inducible promoter were streaked onto SGal-Ura plates and incubated for 5 days at 37 °C. We tested charge-neutralizing (A) and charge-reversal (B) mutations. C, GST pulldown assay of Rpn14 mutants. Affinity-purified GST, GST-Rpn14, GST-Rpn14D157A, and GST-Rpn14D157K were analyzed by SDS-PAGE (left panel). Resin-bound GST or GST fusion proteins were incubated with the E. coli lysate expressing Rpt6-C (+) or not (−). After washing extensively, the resin-bound proteins were eluted and analyzed by SDS-PAGE, followed by silver staining (right panel). WT, wild-type protein; M, molecular mass markers; CBB, Coomassie Brilliant Blue.
Similar articles
- Conformational dynamics of the Rpt6 ATPase in proteasome assembly and Rpn14 binding.
Ehlinger A, Park S, Fahmy A, Lary JW, Cole JL, Finley D, Walters KJ. Ehlinger A, et al. Structure. 2013 May 7;21(5):753-65. doi: 10.1016/j.str.2013.02.021. Epub 2013 Apr 4. Structure. 2013. PMID: 23562395 Free PMC article. - Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p.
Takagi K, Kim S, Yukii H, Ueno M, Morishita R, Endo Y, Kato K, Tanaka K, Saeki Y, Mizushima T. Takagi K, et al. J Biol Chem. 2012 Apr 6;287(15):12172-82. doi: 10.1074/jbc.M112.345876. Epub 2012 Feb 8. J Biol Chem. 2012. PMID: 22334676 Free PMC article. - Chaperone-mediated pathway of proteasome regulatory particle assembly.
Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D. Roelofs J, et al. Nature. 2009 Jun 11;459(7248):861-5. doi: 10.1038/nature08063. Nature. 2009. PMID: 19412159 Free PMC article. - Assembly manual for the proteasome regulatory particle: the first draft.
Park S, Tian G, Roelofs J, Finley D. Park S, et al. Biochem Soc Trans. 2010 Feb;38(Pt 1):6-13. doi: 10.1042/BST0380006. Biochem Soc Trans. 2010. PMID: 20074027 Free PMC article. Review. - Assembly and function of the proteasome.
Saeki Y, Tanaka K. Saeki Y, et al. Methods Mol Biol. 2012;832:315-37. doi: 10.1007/978-1-61779-474-2_22. Methods Mol Biol. 2012. PMID: 22350895 Review.
Cited by
- New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 Å resolution.
Kim S, Nishide A, Saeki Y, Takagi K, Tanaka K, Kato K, Mizushima T. Kim S, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 May 1;68(Pt 5):517-21. doi: 10.1107/S1744309112011359. Epub 2012 Apr 20. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012. PMID: 22691779 Free PMC article. - Conformational dynamics of the Rpt6 ATPase in proteasome assembly and Rpn14 binding.
Ehlinger A, Park S, Fahmy A, Lary JW, Cole JL, Finley D, Walters KJ. Ehlinger A, et al. Structure. 2013 May 7;21(5):753-65. doi: 10.1016/j.str.2013.02.021. Epub 2013 Apr 4. Structure. 2013. PMID: 23562395 Free PMC article. - Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p.
Takagi K, Kim S, Yukii H, Ueno M, Morishita R, Endo Y, Kato K, Tanaka K, Saeki Y, Mizushima T. Takagi K, et al. J Biol Chem. 2012 Apr 6;287(15):12172-82. doi: 10.1074/jbc.M112.345876. Epub 2012 Feb 8. J Biol Chem. 2012. PMID: 22334676 Free PMC article. - Structural biology of the proteasome.
Kish-Trier E, Hill CP. Kish-Trier E, et al. Annu Rev Biophys. 2013;42:29-49. doi: 10.1146/annurev-biophys-083012-130417. Epub 2013 Feb 13. Annu Rev Biophys. 2013. PMID: 23414347 Free PMC article. Review. - Wiggle and Shake: Managing and Exploiting Conformational Dynamics during Proteasome Biogenesis.
Betancourt D, Lawal T, Tomko RJ Jr. Betancourt D, et al. Biomolecules. 2023 Aug 6;13(8):1223. doi: 10.3390/biom13081223. Biomolecules. 2023. PMID: 37627288 Free PMC article. Review.
References
- Coux O., Tanaka K., Goldberg A. L. (1996) Annu. Rev. Biochem. 65, 801–847 - PubMed
- Baumeister W., Walz J., Zühl F., Seemüller E. (1998) Cell 92, 367–380 - PubMed
- Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) Cell 94, 615–623 - PubMed
- Leggett D. S., Glickman M. H., Finley D. (2005) Methods Mol. Biol. 301, 57–70 - PubMed
- Murata S., Yashiroda H., Tanaka K. (2009) Nat. Rev. Mol. Cell Biol. 10, 104–115 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases