Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells - PubMed (original) (raw)
Comparative Study
Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells
Odessa Yabut et al. J Neurosci. 2010.
Abstract
Dyrk1A is a member of the mammalian Dyrk [dual-specificity tyrosine-(Y)-phosphorylation regulated kinase] family of protein kinases that is expressed at high levels in the brain, but its role in the development and function of this organ is not well understood. The human DYRK1A gene is located on trisomic chromosome 21 in Down syndrome (DS) patients, leading to its overexpression. Dyrk1A is also overexpressed in animal models of DS and in gene-specific transgenic mice that consistently exhibit cognitive impairment. To elucidate the cellular and molecular mechanisms that are affected by increased levels of Dyrk1A in the developing brain, we overexpressed this kinase in the embryonic mouse neocortex using the in utero electroporation technique. We found that Dyrk1A overexpression inhibits neural cell proliferation and promotes premature neuronal differentiation in the developing cerebral cortex without affecting cell fate and layer positioning. These effects are dependent on the Dyrk1A kinase activity and are mediated by the nuclear export and degradation of cyclin D1. This study identifies specific Dyrk1A-induced mechanisms that disrupt the normal process of corticogenesis and possibly contribute to cognitive impairment observed in DS patients and animal models.
Figures
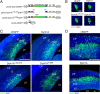
Figure 1.
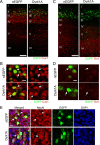
In utero electroporation of Dyrk1A–EGFP constructs in the mouse neocortex. A, Diagram of the plasmids used for IUE. All constructs contained the CAG hybrid promoter, an NLS, and the EGFP reporter fused in-frame to Dyrk1A coding sequences. Expression constructs encode wild-type Dyrk1A (pCAG–Dyrk1AEGFP), a kinase domain deletion (pCAG–Dyrk1AΔ165-478EGFP), and a kinase mutation (pCAG–Dyrk1AK188REGFP). A construct encoding nuclear EGFP alone (pCAG–nEGFP) with an NLS was used as a control. H, Histidine repeat; S/T, serine/threonine-rich regions. B, Subcellular localization of Dyrk1A proteins in transfected neural cells. Confocal images of individual neural cells in the embryonic neocortex expressing Dyrk1A proteins or control nEGFP (green). All proteins accumulated in the nucleus as indicated by colocalization with DAPI (blue), but Dyrk1A proteins exhibited a speckle-like distribution pattern. C, Expression pattern of transfected cells 24 h after IUE. Coronal sections of the lateral neocortex were obtained and imaged by direct EGFP fluorescence (green) and DAPI counterstain (blue) using confocal microscopy. Cells expressing nEGFP as well as Dyrk1AK188R and Dyrk1AΔ165-478 appeared to be widely distributed in the VZ, SVZ, and IZ. Dyrk1A+ cells were primarily excluded from the VZ. D, Expression of transfected cells 48 h after IUE. Representative images of the neocortex were obtained as above, showing that Dyrk1A+ cells were preferentially excluded from the VZ. Scale bars: B, 5 μm; C, D, 500 μm. LV, Lateral ventricle; CP, cortical plate.
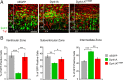
Figure 2.
Neural cells expressing Dyrk1A–EGFP preferentially exit the ventricular zone. A, Embryos were electroporated with the indicated constructs and analyzed 24 h after IUE. Coronal sections of the lateral neocortex were imaged by direct EGFP fluorescence (green) and Tuj1 immunofluorescence (red) using confocal microscopy. LV, Lateral ventricle. Scale bar, 50 μm. B, The percentage of EGFP fluorescent cells in the VZ, SVZ, and IZ was quantified using confocal _z_-stack images collected as in A. The percentage of cells expressing wild-type Dyrk1A was significantly reduced in the VZ and in the SVZ, and it was increased in the IZ compared with control nEGFP or mutant Dyrk1AK188R. Error bars indicate SEM. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
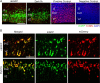
Figure 3.
The absence of Dyrk1A–EGFP cells in the VZ is not attributable to cell death or asymmetric protein localization. A, Dyrk1A overexpression in neural progenitor cells does not induce cell death. nEGFP or Dyrk1A–EGFP constructs were electroporated at E14.5, and TUNEL assay was conducted on E15.5 sections. Virtually no TUNEL staining (red) was observed in nEGFP+ and Dyrk1A+ (green) cells. DNase-treated sections were used as a positive control. As a negative control, sections were not treated with terminal deoxyribonucleotide transferase enzyme. Control sections are shown with DAPI counterstaining (blue). Scale bar, 100 μm. B, Coexpression of mCherry and Dyrk1A–EGFP or control nEGFP. Coronal sections of the medial neocortex were imaged to visualize EGFP and mCherry. The distribution of the mCherry signal was similar to that of EGFP for all electroporated constructs. Scale bar, 50 μm.
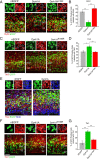
Figure 4.
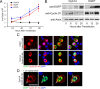
Dyrk1A overexpression inhibits cell cycle progression and induces premature neuronal differentiation in a kinase-dependent manner. E14.5 mouse embryos were electroporated in utero with the indicated plasmids. Representative confocal images were obtained from the lateral neocortex 24 h after IUE. Sections were imaged by EGFP autofluorescence (green) to visualize the transfected proteins, immunofluorescence for the indicated antibodies (red), and DAPI staining (blue). Higher magnifications of the boxed areas are shown above each overlay image to illustrate representative patterns of colocalization. A, BrdU was injected in the pregnant dam 4 h before killing the embryos to identify proliferating cells. B, Quantification of the BrdU colabeling data. The percentage of cells expressing wild-type Dyrk1A that were positive for BrdU was significantly reduced compared with cells expressing control nEGFP or mutant Dyrk1AK188R. C, Coexpression of the intermediate progenitor marker Tbr2. D, Quantification of Tbr2 coexpression data. The percentage of Dyrk1A+ cells that coexpressed Tbr2 was significantly higher compared with nEGFP+ and mutant Dyrk1AK188R+ cells. E, BrdU labeling of Tbr2+ cells. F, Coexpression of the early postmitotic neuronal marker Tbr1. G, Quantification of the Tbr1 coexpression data. The percentage of cells expressing wild-type Dyrk1A that was positive for Tbr1 was significantly increased compared with cells expressing control nEGFP+ and mutant Dyrk1AK188R. Scale bars, 50 μm. Bar graphs show SEM. *p ≤ 0.05; **p ≤ 0.01.
Figure 5.
Dyrk1A-overexpressing cells migrated and differentiated normally in the postnatal neocortex. Postnatal day 6 brains were obtained from pups electroporated at E14.5 with plasmids encoding Dyrk1A–EGFP or nEGFP. A–E, Coronal sections from the lateral neocortex were subjected to immunofluorescence staining against the upper layer marker Cux1 (A, B), the deep layer marker Tbr1 (C, D), or the mature neuronal marker NeuN (E). Confocal _z-_stack images show that Dyrk1A+ and nEGFP+ cells (green) were positioned in the upper cortical layers, expressed Cux1 but not Tbr1, and also expressed the mature neuronal marker NeuN. Scale bars: A, C, 100 μm; B, D, E, 25 μm.
Figure 6.
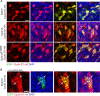
Dyrk1A expression in Neuro2a cells causes proliferation arrest and cyclin D1 nuclear export. A, Neuro2a cells expressing Dyrk1A failed to proliferate. Cells were transfected with pDyrk1A–EGFP or pEGFP, and the number of fluorescent cells was determined in triplicate samples using a hemocytometer. Untransfected (EGFP−) cells from the same cultures were also counted. Line graphs show the SEM. B, Dyrk1A expression induces cyclin D1 degradation. Neuro2a cells were transfected with pDyrk1A–EGFP or pEGFP plasmids and lysed at 0, 6, 12, and 20 h after transfection. Western blot analysis was conducted using antibodies against cyclin D1, EGFP, and actin as a loading control. C, Neuro2a cells cotransfected with plasmids encoding HA-tagged cyclin D1 and EGFP (top row) or Dyrk1A–EGFP (bottom row) were subjected to immunofluorescence staining using anti-HA antibodies. Confocal _z_-stack images were obtained to analyze the subcellular localization of HA-labeled cyclin D1 (red) in EGFP+ cells (green). Cyclin D1 was predominantly nuclear in cells transfected with pEGFP but appeared to be mostly cytoplasmic in cells transfected with pDyrk1A–EGFP. Scale bar, 25 μm. D, Dyrk1A and cyclin D1 colocalized in nuclear aggregates in transfected Neuro2a cells. High-resolution confocal images of a single 1 μm optical section showing the colocalization of Dyrk1A–EGFP and cyclin D1–HA in many nuclear aggregates near the nuclear membrane. Scale bar, 10 μm.
Figure 7.
Dyrk1A overexpression in neural cells in vivo promotes cyclin D1 nuclear export. E14.5 mouse embryos were coelectroporated with pCAG–cyclin D1–HA and pCAG–nEGFP, pCAG–Dyrk1AEGFP, or pCAG–Dyrk1AK188REGFP. After 24 h, coronal sections of the lateral neocortex were obtained, and confocal _z_-stack images were acquired to visualize EGFP expression (green) and cyclin D1–HA localization (red). Sections were counterstained with the nuclear marker DAPI (blue). A, Cyclin D1 appeared to be mostly nuclear in nEGFP+ or Dyrk1AK188R+ cells. Cells expressing Dyrk1A exhibited either mostly cytoplasmic cyclin D1 or nuclear aggregates containing the cyclin. Scale bar, 10 μm. B, High-resolution confocal images of a single 1 μm optical section showing the colocalization of Dyrk1A–EGFP and cyclin D1 in nuclear aggregates. Scale bar, 5 μm.
Figure 8.
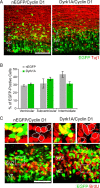
Cyclin D1 rescued the proliferation defect induced by Dyrk1A overexpression in vivo. E14.5 embryos were coelectroporated with pCAG–cyclin D1–HA and pCAG–nEGFP or pCAG–Dyrk1AEGFP and pulsed by BrdU injection. At 24 h after IUE, coronal sections of the lateral neocortex were subjected to immunofluorescence staining against Tuj1 (A) or BrdU (C). A, Representative confocal images showing the localization of transfected cells (green) and Tuj1 immunofluorescence (red). B, Quantification of the data obtained as in A. The localization of cells expressing Dyrk1A in different zones of the neocortex was comparable with that of control nEGFP-expressing cells. C, BrdU labeling of embryos coelectroporated in utero with cyclin D1 and nEGFP or Dyrk1A plasmids. Higher magnifications of the boxed areas are shown above each overlay image to illustrate representative patterns of colocalization. The percentage of cells expressing Dyrk1A that was positive for BrdU did not differ significantly when compared with control nEGFP+ cells. Scale bars: A, 50 μm; C, 100 μm. Bar graphs show SEM.
Similar articles
- Overexpression of DYRK1A inhibits choline acetyltransferase induction by oleic acid in cellular models of Down syndrome.
Hijazi M, Fillat C, Medina JM, Velasco A. Hijazi M, et al. Exp Neurol. 2013 Jan;239:229-34. doi: 10.1016/j.expneurol.2012.10.016. Epub 2012 Nov 1. Exp Neurol. 2013. PMID: 23124096 - Effect of DYRK1A activity inhibition on development of neuronal progenitors isolated from Ts65Dn mice.
Mazur-Kolecka B, Golabek A, Kida E, Rabe A, Hwang YW, Adayev T, Wegiel J, Flory M, Kaczmarski W, Marchi E, Frackowiak J. Mazur-Kolecka B, et al. J Neurosci Res. 2012 May;90(5):999-1010. doi: 10.1002/jnr.23007. Epub 2012 Jan 18. J Neurosci Res. 2012. PMID: 22252917 - Engineering DYRK1A overdosage yields Down syndrome-characteristic cortical splicing aberrations.
Toiber D, Azkona G, Ben-Ari S, Torán N, Soreq H, Dierssen M. Toiber D, et al. Neurobiol Dis. 2010 Oct;40(1):348-59. doi: 10.1016/j.nbd.2010.06.011. Epub 2010 Jun 30. Neurobiol Dis. 2010. PMID: 20600907 - [Molecular Mechanism Underlying Abnormal Differentiation of Neural Progenitor Cells in the Developing Down Syndrome Brain].
Kurabayashi N, Sanada K. Kurabayashi N, et al. Yakugaku Zasshi. 2017;137(7):795-800. doi: 10.1248/yakushi.16-00236-1. Yakugaku Zasshi. 2017. PMID: 28674289 Review. Japanese. - DYRK1A (dual-specificity tyrosine-phosphorylated and -regulated kinase 1A): a gene with dosage effect during development and neurogenesis.
Dierssen M, de Lagrán MM. Dierssen M, et al. ScientificWorldJournal. 2006 Jun 17;6:1911-22. doi: 10.1100/tsw.2006.319. ScientificWorldJournal. 2006. PMID: 17205196 Free PMC article. Review.
Cited by
- E3 Ligase SCFβTrCP-induced DYRK1A Protein Degradation Is Essential for Cell Cycle Progression in HEK293 Cells.
Liu Q, Tang Y, Chen L, Liu N, Lang F, Liu H, Wang P, Sun X. Liu Q, et al. J Biol Chem. 2016 Dec 16;291(51):26399-26409. doi: 10.1074/jbc.M116.717553. Epub 2016 Nov 2. J Biol Chem. 2016. PMID: 27807027 Free PMC article. - Dyrk1a Mutations Cause Undergrowth of Cortical Pyramidal Neurons via Dysregulated Growth Factor Signaling.
Levy JA, LaFlamme CW, Tsaprailis G, Crynen G, Page DT. Levy JA, et al. Biol Psychiatry. 2021 Sep 1;90(5):295-306. doi: 10.1016/j.biopsych.2021.01.012. Epub 2021 Apr 8. Biol Psychiatry. 2021. PMID: 33840455 Free PMC article. - Cyclin D1 Again Caught in the Act: Dyrk1a Links G1 and Neurogenesis in Down Syndrome.
Smith I, Calegari F. Smith I, et al. EBioMedicine. 2015 Feb 7;2(2):96-7. doi: 10.1016/j.ebiom.2015.02.003. eCollection 2015. EBioMedicine. 2015. PMID: 26137545 Free PMC article. Review. No abstract available. - Meeting at the crossroads: common mechanisms in Fragile X and Down syndrome.
Chang KT, Ro H, Wang W, Min KT. Chang KT, et al. Trends Neurosci. 2013 Dec;36(12):685-94. doi: 10.1016/j.tins.2013.08.007. Epub 2013 Sep 25. Trends Neurosci. 2013. PMID: 24075449 Free PMC article. Review. - An atypical DYRK kinase connects quorum-sensing with posttranscriptional gene regulation in Trypanosoma brucei.
Cayla M, McDonald L, MacGregor P, Matthews K. Cayla M, et al. Elife. 2020 Mar 26;9:e51620. doi: 10.7554/eLife.51620. Elife. 2020. PMID: 32213288 Free PMC article.
References
- Ahn KJ, Jeong HK, Choi HS, Ryoo SR, Kim YJ, Goo JS, Choi SY, Han JS, Ha I, Song WJ. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol Dis. 2006;22:463–472. - PubMed
- Altafaj X, Dierssen M, Baamonde C, Martí E, Visa J, Guimerà J, Oset M, González JR, Flórez J, Fillat C, Estivill X. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum Mol Genet. 2001;10:1915–1923. - PubMed
- Alvarez M, Estivill X, de la Luna S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J Cell Sci. 2003;116:3099–3107. - PubMed
- Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials