Dendritic cells produce CXCL13 and participate in the development of murine small intestine lymphoid tissues - PubMed (original) (raw)
Dendritic cells produce CXCL13 and participate in the development of murine small intestine lymphoid tissues
Keely G McDonald et al. Am J Pathol. 2010 May.
Abstract
In the adult intestine, luminal microbiota induce cryptopatches to transform into isolated lymphoid follicles (ILFs), which subsequently act as sites for the generation of IgA responses. The events leading to this conversion are incompletely understood. Dendritic cells (DCs) are components of cryptopatches (CPs) and ILFs and were therefore evaluated in this process. We observed that the adult murine intestine contains clusters of DCs restricted to the CP/ILF continuum. A numerical and cell associative hierarchy in the adult intestine and a chronologic hierarchy in the neonatal intestine demonstrated that these clusters form after the coalescence of CD90+ cells to form CPs and before the influx of B220+ B lymphocytes to form ILFs. Cluster formation was dependent on lymphotoxin and the lymphotoxin beta receptor and independent of lymphocytes. The ILF DC population was distinguished from that of the lamina propria by the absence of CD4+CD11c+ cells and an increased proportion of CD11c+B220+ cells. The formation of clusters was not limited by DC numbers but was induced by luminal microbiota. Moreover, in the absence of the chemokine CXCL13, CP transformation into ILF was arrested. Furthermore, ILF DCs express CXCL13, and depletion of DCs resulted in regression of ILFs and disorganization of CPs. These results reveal DC participation in ILF transformation and maintenance and suggest that in part this may be due to CXCL13 production by these cells.
Figures
Figure 1
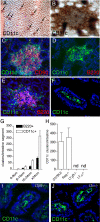
Intestinal DCs form discreet clusters that are part of the CP/ILF continuum. Using anti-CD11c-stained whole mounts (brown staining, A and B) and traditional immunohistochemistry (green staining, C–F, I, and J), we evaluated the regional location, organization, and cellular associations intestinal DCs in wild-type (A–G) and genetically manipulated mice on the C57BL/6 background (H–J). We observed that the intestine contained multiple clusters of DCs (arrows, A), and these clusters were located at the base of villi (en face view of whole mount at higher power, B). All of the DC clusters we observed were associated with CD90+ cells (C), consistent with their localization to CP. The DC clusters contained variable numbers of B220+ cells; some containing no B220+ cells (D) and others colocalizing with B220+ clusters (E). DC clusters were more numerous than B220+ clusters but had an identical regional distribution (G). Like the formation of ILFs and CPs, the formation of DC clusters was dependent on LT and the LTβR but independent of the presence of T or B lymphocytes (H). However unlike the development of DC clusters, the presence of LP DCs was not dependent on LTα or LTβR (I and J). n = 3 or more animals from each group for data in G and H. nd = none detected. Scales bars in panels A = 500 μm and in panels B and C = 100 μm.
Figure 2
CD11c+ clusters develop after CD90+ clusters and before B220+ clusters in the neonatal intestine. CPs and ILFs first appear in the neonatal period. To examine the chronology of cellular cluster formation in SILT development, we evaluated the presence of CD90+, CD11c+, or B220+ clusters in the intestine of C57BL/6 neonatal mice. None of these cellular clusters was present before day 17 of life (A). CD90+ clusters were present on day 17 of life, a few of these clusters contained CD11c+ cells, none of these clusters contained B220+ cells (A–D). On day 18 of life, CD11c+ clusters were present and were nearly as numerous as the CD90+ clusters (A). In each instance the CD11c+ clusters were associated with CD90+ clusters, and a few clusters contained B220+ cells (E and F). The full complement of cellular clusters was present by day 19 of life and in proportions that approximate that seen in the adult intestine (A). All CD90+ clusters were also c-kit+ (G). Although DC clusters were rare, LP DC populations were readily apparent on day 17 of neonatal life (H). n = 3 or more mice for each time point and type of cellular cluster in examined in A. Scale bar = 50 μm.
Figure 3
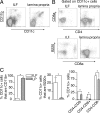
ILF and LP CD11c+ cell populations have different phenotypes. To compare the phenotype of CD11c+ cells located within the clusters to those located within the general LP, cellular populations were isolated from ILFs and LP and examined with multicolor flow cytometry. ILF CD11c+ cells were largely CD11b−, whereas in comparison those from the LP were largely CD11b+ (A and C). The ILFs contained almost no CD4+CD11c+ cells, whereas this cellular population was prominent in the LP (B and C). Conversely CD11c+B220+ and CD11c+CD8α+ cells were relatively enriched in ILFs when compared with the LP (B and C). Using CD4 and CD8α expression to classify the CD11c+ cell subtypes, we observed that CD11c+ cellular population from both the LP and ILFs was predominantly CD4−CD8α−, whereas the LP population was relatively enriched in CD4+CD8α− cells, and the ILF population was relatively enriched in CD4−CD8α+ cells (C). n = 3 or more replicates using ILF cellular populations pooled from multiple animals and LP populations for data in C. *P < 0.05.
Figure 4
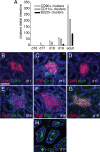
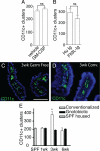
DC numbers are not limiting for the development of CD11c+ clusters. Cluster formation can be driven by changes in luminal microbiota. To assess the availability of DCs as a limiting factor in the development of DC clusters, we treated C57BL/6 mice for 2 weeks with exogenous GM-CSF or Flt3L-Ig to increase DC numbers and then evaluated the numbers of CD11c+ clusters in the intestine. Exogenous GM-CSF or Flt3L-Ig had no effect on the number of CD11c+ clusters (A and B). To evaluate the role of luminal microbiota in CD11c+ cluster formation, the presence of LP DC populations and the number of clusters in the distal one-third of the intestine from gnotobiotic mice that remained germfree or their counterparts that were given cecal contents from conventionally housed C57BL/6 mice were examined. Three weeks following conventionalization, the presence of the LP DC population appeared similar in germfree mice and their conventionalized counterparts (C and D). The numbers of CD11c+ clusters in the mice that remained germfree throughout the experiment did not change, and this was similar to the number of clusters seen in the intestine of conventionally housed C57BL/6 mice (E). Conversely the numbers of CD11c+ clusters increased significantly 3 weeks following colonization and returned to levels seen in specific pathogen-free housed animals germ free animals 3 weeks later (E). n = 3 or more mice in each group and each time point. *P < 0.05. Scale bar = 100 μm.
Figure 5
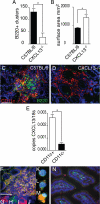
In the absence of CXCL13, ILF development is arrested and abnormal, and ILF CD11c+ cells are sources of CXCL13. Intestines from wild-type C57BL/6 mice and CXCL13−/− mice on the C57BL/6 background were evaluated for the presence of ILFs (B220+ clusters, A) and the size (B) and morphology of the CD11c+ clusters (C and D). CXCL13−/− mice were found to have significantly fewer ILFs (A), and conversely the surface area of the B220−CD11c+ cellular clusters was found to be significantly greater in CXCL13−/− mice (B). The CD11c+B220− clusters in the CXCL13−/− intestine were of a similar size to CD11c+B220+ clusters from wild-type mice but did not contain a central area filled with mononuclear cells and instead extended to engulf multiple adjacent crypts (C and D). CD11c+ and CD11c− cellular populations were isolated from ILFs from C57BL/6 mice as described in Materials and Methods, and the mRNA expression of CXCL13 was examined. CD11c+ ILF cells were found to express significantly more CXCL13 than CD11c− ILF cells (E). CXCL13 protein expression was evaluated using immunohistochemistry. CXCL13 protein (red staining, F) was detected within ILFs in the intestine of C5BL/6 mice. Many of the CXCL13-expressing cells were also CD11c+ (green staining, F; costaining appears yellow). Insets (G and H) demonstrate individual channels for cells within ILFs costaining for both CXCL13 (red, G) and CD11c (green, H). Insets (I and J) demonstrate individual channels for cells within ILFs staining for CXCL13 (red, I) and not staining for CD11c (lack of green signal, J). To confirm the results seen in whole tissues, we evaluated the protein expression of CXCL13 in isolated ILF cellular populations. ILF cells expressed CXCL13 (red signal, M) and that these cells could be CD11c+ (green signal, M; costaining appears yellow). We also observed that some CD11c+ cells (green signal, L) did not express CXCL13 (lack of red signal, L), and some cells were neither CD11c+ or CXCL13+ (lack of red and green signal, K). We did not observe CXCL13 expression by DCs located within the LP (N green = CD11c, red = CXCL13). n = 3 intestines A and B. E is representative of one of two independent experiments using pooled ILF cellular populations from three or more animals. Scale bar = 100 μm. *P < 0.05.
Figure 6
DCs participate in the maintenance of B lymphocytes in ILFs. To evaluate a role for CD11c+ cells in ILF maintenance CD11c-DTR transgenic mice (Tg) and wild-type C57BL/6 mice (WT) were injected with DT i.p. and evaluated for the presence of ILFs (B220+ clusters, A) and the morphology and the presence of CD90+, CD11c+, and B220+ cells in the SILT in the Tg animals (B and C). Following DT treatment, B220+ clusters were maintained in WT but were significantly decreased in the transgenic animals (A). CD90+ clusters could be easily identified in the Tg following DT treatment; however, these clusters lacked CD11c+ and B220+ cells were disorganized and engulfed the adjacent crypts (B and C). n = 3 or more animals for each condition. *P < 0.05 when compared with the WT treatment group in A. Scale bar = 100 μm.
Similar articles
- Distinct developmental requirements for isolated lymphoid follicle formation in the small and large intestine: RANKL is essential only in the small intestine.
Knoop KA, Butler BR, Kumar N, Newberry RD, Williams IR. Knoop KA, et al. Am J Pathol. 2011 Oct;179(4):1861-71. doi: 10.1016/j.ajpath.2011.06.004. Epub 2011 Aug 18. Am J Pathol. 2011. PMID: 21854748 Free PMC article. - Isolated lymphoid follicle formation is inducible and dependent upon lymphotoxin-sufficient B lymphocytes, lymphotoxin beta receptor, and TNF receptor I function.
Lorenz RG, Chaplin DD, McDonald KG, McDonough JS, Newberry RD. Lorenz RG, et al. J Immunol. 2003 Jun 1;170(11):5475-82. doi: 10.4049/jimmunol.170.11.5475. J Immunol. 2003. PMID: 12759424 - Induction of intestinal lymphoid tissue: the role of cryptopatches.
Lügering A, Kucharzik T. Lügering A, et al. Ann N Y Acad Sci. 2006 Aug;1072:210-7. doi: 10.1196/annals.1326.015. Ann N Y Acad Sci. 2006. PMID: 17057201 Review. - CCR6 and CCL20: partners in intestinal immunity and lymphorganogenesis.
Williams IR. Williams IR. Ann N Y Acad Sci. 2006 Aug;1072:52-61. doi: 10.1196/annals.1326.036. Ann N Y Acad Sci. 2006. PMID: 17057190 Review.
Cited by
- Intestinal Antigen-Presenting Cells: Key Regulators of Immune Homeostasis and Inflammation.
Flannigan KL, Geem D, Harusato A, Denning TL. Flannigan KL, et al. Am J Pathol. 2015 Jul;185(7):1809-19. doi: 10.1016/j.ajpath.2015.02.024. Epub 2015 May 11. Am J Pathol. 2015. PMID: 25976247 Free PMC article. Review. - Immune Profiling of Human Gut-Associated Lymphoid Tissue Identifies a Role for Isolated Lymphoid Follicles in Priming of Region-Specific Immunity.
Fenton TM, Jørgensen PB, Niss K, Rubin SJS, Mörbe UM, Riis LB, Da Silva C, Plumb A, Vandamme J, Jakobsen HL, Brunak S, Habtezion A, Nielsen OH, Johansson-Lindbom B, Agace WW. Fenton TM, et al. Immunity. 2020 Mar 17;52(3):557-570.e6. doi: 10.1016/j.immuni.2020.02.001. Epub 2020 Mar 10. Immunity. 2020. PMID: 32160523 Free PMC article. - Differential Effect of Light and Dark Period Sleep Fragmentation on Composition of Gut Microbiome and Inflammation in Mice.
Sanford LD, Wellman LL, Ciavarra RP, Oldfield EC 4th, Shams R, Copare JL, Johnson DA. Sanford LD, et al. Life (Basel). 2021 Nov 23;11(12):1283. doi: 10.3390/life11121283. Life (Basel). 2021. PMID: 34947814 Free PMC article. - Oestrogen-mediated protection of experimental autoimmune encephalomyelitis in the absence of Foxp3+ regulatory T cells implicates compensatory pathways including regulatory B cells.
Subramanian S, Yates M, Vandenbark AA, Offner H. Subramanian S, et al. Immunology. 2011 Mar;132(3):340-7. doi: 10.1111/j.1365-2567.2010.03380.x. Epub 2010 Nov 23. Immunology. 2011. PMID: 21091909 Free PMC article. - Distinct developmental requirements for isolated lymphoid follicle formation in the small and large intestine: RANKL is essential only in the small intestine.
Knoop KA, Butler BR, Kumar N, Newberry RD, Williams IR. Knoop KA, et al. Am J Pathol. 2011 Oct;179(4):1861-71. doi: 10.1016/j.ajpath.2011.06.004. Epub 2011 Aug 18. Am J Pathol. 2011. PMID: 21854748 Free PMC article.
References
- Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol. 2008;1:11–22. - PubMed
- Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II, Itoh K, Littman DR, Fagarasan S. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–271. - PubMed
- Lorenz RG, Newberry RD. Isolated lymphoid follicles can function as sites for induction of mucosal immune responses. Ann NY Acad Sci. 2004;1029:44–57. - PubMed
- Herbrand H, Bernhardt G, Forster R, Pabst O. Dynamics and function of solitary intestinal lymphoid tissue. Crit Rev Immunol. 2008;28:1–13. - PubMed
- Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK064798/DK/NIDDK NIH HHS/United States
- P30 CA91842/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- AG028309/AG/NIA NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- R21 AG028309/AG/NIA NIH HHS/United States
- DK64798/DK/NIDDK NIH HHS/United States
- P30-DK52574/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous