Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer's Disease and Down syndrome - PubMed (original) (raw)
Truncated beta-amyloid peptide channels provide an alternative mechanism for Alzheimer's Disease and Down syndrome
Hyunbum Jang et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Full-length amyloid beta peptides (Abeta(1-40/42)) form neuritic amyloid plaques in Alzheimer's disease (AD) patients and are implicated in AD pathology. However, recent transgenic animal models cast doubt on their direct role in AD pathology. Nonamyloidogenic truncated amyloid-beta fragments (Abeta(11-42) and Abeta(17-42)) are also found in amyloid plaques of AD and in the preamyloid lesions of Down syndrome, a model system for early-onset AD study. Very little is known about the structure and activity of these smaller peptides, although they could be the primary AD and Down syndrome pathological agents. Using complementary techniques of molecular dynamics simulations, atomic force microscopy, channel conductance measurements, calcium imaging, neuritic degeneration, and cell death assays, we show that nonamyloidogenic Abeta(9-42) and Abeta(17-42) peptides form ion channels with loosely attached subunits and elicit single-channel conductances. The subunits appear mobile, suggesting insertion of small oligomers, followed by dynamic channel assembly and dissociation. These channels allow calcium uptake in amyloid precursor protein-deficient cells. The channel mediated calcium uptake induces neurite degeneration in human cortical neurons. Channel conductance, calcium uptake, and neurite degeneration are selectively inhibited by zinc, a blocker of amyloid ion channel activity. Thus, truncated Abeta fragments could account for undefined roles played by full length Abetas and provide a unique mechanism of AD and Down syndrome pathologies. The toxicity of nonamyloidogenic peptides via an ion channel mechanism necessitates a reevaluation of the current therapeutic approaches targeting the nonamyloidogenic pathway as avenue for AD treatment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
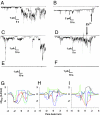
Aβ channel conformations by MD simulations. The simulated channel structures (Left) with highlighted subunits for the N9 (A), p3 (B), and p3-F19P mutant (C) channels are shown in the view along the membrane normal. (Center and Right) Averaged pore structures calculated by the HOLE program (50) embedded in the averaged channel conformations during the simulations. In the angle views of the pore structure (Center), whole channel structures are shown with the ribbon representation. In the lateral views of the pore structure (Right), cross-sectioned channels are given in the surface representation. For the pore structures in the surface representation, the degree of the pore diameter is indicated by the color codes in the order of red < green < blue. In the channel structures, hydrophobic residues are shown in white, polar, and Gly residues are shown in green, positively charged residues are shown in blue, and negatively charged residues are shown in red.
Fig. 2.
AFM imaging. Error-mode AFM images of N9 (A), p3 (B), and p3-F19P mutant (C) reconstituted in lipid bilayers. Individual channels are enclosed by dotted circles and the white arrow in A indicates a mica region free of bilayer. High-resolution images are of individual channel structures. The number of subunits is indicated for each channel. Inner-pore sizes are typically 1 to 2 nm. Images sizes are 22 nm (D), 19 nm (E), 15 nm (F), 23 nm (G), 11 nm (H), and 9 nm (I).
Fig. 3.
Pore-forming activity of nonamyloidogenic peptides. Channel conductance measurements (A_–_F) and potential of mean forces (PMFs) for Mg2+, K+, Ca2+, Zn2+, and Cl− (G to H). Single channel currents induced by N9 (A) and p3 (C). Current jumps on traces correspond to single opening or closing of ion channels. Multilevel conductances can be observed. Blockade of N9 channels (B) and p3 channels (D) by 1 mM ZnCl2. Zinc addition is indicated by arrows. As a control, p3 scrambled sequence shows no membrane activity for extended periods of time (E). As predicted, structurally blocked p3 mutant F19P does not form conductive channels for extended periods of time (F). Planar lipid bilayers were made in electrolyte with 100 mM KCI, 10 mM hepes-K pH 7.4, and 1 mM MgCI2, in all of the experiments. The bilayer membranes shown were made with asolectin lipids (soybean lecithin). The cis side is the virtual ground. The applied membrane potential was −50 mV. The current traces shown are representative of at least 7 and often more than 10 independent experiments for each condition shown. PMF for each ion are shown for N9 (G), p3 (H), and p3-F19P mutant (I) channels. Both N9 and p3 channels preserve the pore; p3-F19P channel has a collapsed pore. Δ_G_PMF, was calculated using the equation  , where _k_B is the Boltzmann constant, T is the simulation temperature, ρz is the density of ion at the position z along the pore axis, and ρbulk is the density of ion in the bulk region, representing the relative free-energy profile for Mg2+ (green lines), K+ (red lines), Ca2+ (blue lines), Zn2+ (cyan lines), and Cl− (black lines) as a function of the distance along the pore center axis of each channel.
, where _k_B is the Boltzmann constant, T is the simulation temperature, ρz is the density of ion at the position z along the pore axis, and ρbulk is the density of ion in the bulk region, representing the relative free-energy profile for Mg2+ (green lines), K+ (red lines), Ca2+ (blue lines), Zn2+ (cyan lines), and Cl− (black lines) as a function of the distance along the pore center axis of each channel.
Fig. 4.
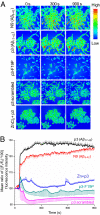
Cell calcium imaging and relative concentration plot. (A) Nonamyloidogenic β-peptides form Ca2+ permeable, zinc-sensitive pores. Time-course measurement of Ca2+ change shows significant variation in the Ca2+ flux upon application of 5 μM N9 and p3 (first and second row, respectively). No increase in Ca2+ was seen in proline mutation and scrambled p3 peptides (third and fourth row, respectively). Pretreatment with zinc, a known Aβ-blocker, prevents such rise in intracellular Ca2+ (fifth row). (B) The plot summarizes the above findings for the entire span of the experiment in all of the cells under the field-of-view. The mean (average of Ca2+ changes in the field) ratio [(Ft/F0)*100] fluorescence intensity change was plotted against time. Ft is the fluorescence of the field at a given time t and F0 is the fluorescence of the field at time t = 0. Error bar represents the SE of the mean of all of the blobs in the field and they are represented on any one side of the curve.
Fig. 5.
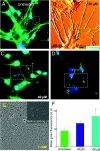
p3-induced dose-dependent neurite degeneration and cell death. Multimodal imaging of cells and processes: immunofluorescence imaging of microtubules (A, C, D), AFM imaging (B), Calcein AM dye (E), and apoptosis assay (F). After 24 h of incubation, 20 μM p3 induced a small neuritic damage only visible in AFM images (B) and ≥40 μM p3 induced loss of neurite density visible by light microscopy [white dotted lines in C (40 μM) and D (100 μM)]. On the other hand, 1 mM of p3 induced rapid neuritic degeneration within 15 min of incubation (E). Significantly, pretreatment with Zn2+ prevented p3-induced damage, even at such a large p3 dose (
Fig. S5
). The original fluorescence images (
Fig. S5
) were processed to reveal the neurites that were obscured by the leakage of Calcein dye. Laplacian of Gaussian (LoG) operator was applied on the image to detect the neurites faithfully. The bar chart represents mean fluorescence (see
SI Materials and Methods
for details) of apoptotic cells (F). Compared with untreated population, 100 μM p3 causes significant (*P < 0.01) cell damage within 24 h of incubation.
Similar articles
- Atomic force microscopy and MD simulations reveal pore-like structures of all-D-enantiomer of Alzheimer's β-amyloid peptide: relevance to the ion channel mechanism of AD pathology.
Connelly L, Jang H, Arce FT, Capone R, Kotler SA, Ramachandran S, Kagan BL, Nussinov R, Lal R. Connelly L, et al. J Phys Chem B. 2012 Feb 9;116(5):1728-35. doi: 10.1021/jp2108126. Epub 2012 Jan 25. J Phys Chem B. 2012. PMID: 22217000 Free PMC article. - The "nonamyloidogenic" p3 fragment (amyloid beta17-42) is a major constituent of Down's syndrome cerebellar preamyloid.
Lalowski M, Golabek A, Lemere CA, Selkoe DJ, Wisniewski HM, Beavis RC, Frangione B, Wisniewski T. Lalowski M, et al. J Biol Chem. 1996 Dec 27;271(52):33623-31. doi: 10.1074/jbc.271.52.33623. J Biol Chem. 1996. PMID: 8969231 - Characterization of monomeric and soluble aggregated Aβ in Down's syndrome and Alzheimer's disease brains.
Gkanatsiou E, Sahlin C, Portelius E, Johannesson M, Söderberg L, Fälting J, Basun H, Möller C, Odergren T, Zetterberg H, Blennow K, Lannfelt L, Brinkmalm G. Gkanatsiou E, et al. Neurosci Lett. 2021 May 29;754:135894. doi: 10.1016/j.neulet.2021.135894. Epub 2021 Apr 10. Neurosci Lett. 2021. PMID: 33848613 - Alzheimer's disease.
De-Paula VJ, Radanovic M, Diniz BS, Forlenza OV. De-Paula VJ, et al. Subcell Biochem. 2012;65:329-52. doi: 10.1007/978-94-007-5416-4_14. Subcell Biochem. 2012. PMID: 23225010 Review. - Amyloid beta ion channel: 3D structure and relevance to amyloid channel paradigm.
Lal R, Lin H, Quist AP. Lal R, et al. Biochim Biophys Acta. 2007 Aug;1768(8):1966-75. doi: 10.1016/j.bbamem.2007.04.021. Epub 2007 May 3. Biochim Biophys Acta. 2007. PMID: 17553456 Free PMC article. Review.
Cited by
- Aggregation Propensities of Herpes Simplex Virus-1 Proteins and Derived Peptides: An In Silico and In Vitro Analysis.
Singh VK, Kumar S, Tapryal S. Singh VK, et al. ACS Omega. 2020 May 26;5(22):12964-12973. doi: 10.1021/acsomega.0c00730. eCollection 2020 Jun 9. ACS Omega. 2020. PMID: 32548480 Free PMC article. - The structure of tyrosine-10 favors ionic conductance of Alzheimer's disease-associated full-length amyloid-β channels.
Karkisaval AG, Hassan R, Nguyen A, Balster B, Abedin F, Lal R, Tatulian SA. Karkisaval AG, et al. Nat Commun. 2024 Feb 13;15(1):1296. doi: 10.1038/s41467-023-43821-y. Nat Commun. 2024. PMID: 38351257 Free PMC article. - Nanoscale electrostatic domains in cholesterol-laden lipid membranes create a target for amyloid binding.
Drolle E, Gaikwad RM, Leonenko Z. Drolle E, et al. Biophys J. 2012 Aug 22;103(4):L27-9. doi: 10.1016/j.bpj.2012.06.053. Biophys J. 2012. PMID: 22947946 Free PMC article. - Simple, Reliable Protocol for High-Yield Solubilization of Seedless Amyloid-β Monomer.
Taylor AIP, Davis PJ, Aubrey LD, White JBR, Parton ZN, Staniforth RA. Taylor AIP, et al. ACS Chem Neurosci. 2023 Jan 4;14(1):53-71. doi: 10.1021/acschemneuro.2c00411. Epub 2022 Dec 13. ACS Chem Neurosci. 2023. PMID: 36512740 Free PMC article. - Imaging Amyloid-β Membrane Interactions: Ion-Channel Pores and Lipid-Bilayer Permeability in Alzheimer's Disease.
Viles JH. Viles JH. Angew Chem Int Ed Engl. 2023 Jun 19;62(25):e202215785. doi: 10.1002/anie.202215785. Epub 2023 Mar 30. Angew Chem Int Ed Engl. 2023. PMID: 36876912 Free PMC article. Review.
References
- Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. - PubMed
- Walsh DM, et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. - PubMed
- Cheng IH, et al. Accelerating amyloid-β fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J Biol Chem. 2007;282:23818–23828. - PubMed
- Kagan BL. Amyloidosis and protein folding. Science. 2005;307:42–43. author reply 42–43. - PubMed
- Jang H, Zheng J, Lal R, Nussinov R. New structures help the modeling of toxic amyloidbeta ion channels. Trends Biochem Sci. 2008;33:91–100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous