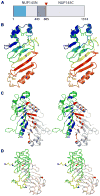Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145 - PubMed (original) (raw)
Sinem A Ozyurt, Johnny Do, Kevin T Bain, Mark Dickey, Logan A Rodgers, Tarun Gheyi, Andrej Sali, Seung Joong Kim, Jeremy Phillips, Ursula Pieper, Javier Fernandez-Martinez, Josef D Franke, Anne Martel, Hiro Tsuruta, Shane Atwell, Devon A Thompson, J Spencer Emtage, Stephen R Wasserman, Michael P Rout, J Michael Sauder, Stephen K Burley
Affiliations
- PMID: 20310066
- PMCID: PMC3136511
- DOI: 10.1002/prot.22707
Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145
Parthasarathy Sampathkumar et al. Proteins. 2010 Jun.
No abstract available
Figures
Figure 1
A: Schematic representation of the S. cerevisiae Nup145 protein. The “FG” repeats are shaded in blue and the expression construct boundaries of Nup145N(443–605) delimited in gray. The site of autoproteolysis is marked with a red triangle. B: Stereoview of the Nup145N(443–605) monomer. Cartoon of Chain B is shown as a rainbow from blue to red from N- to C-terminus. C: Stereoview of the Nup145N(443–605) head-to-tail dimer seen in the tetragonal crystals. Cartoons of Chains A and B are shown in grey and green, respectively. Residues involved in dimeric interactions from Chain B are colored in blue and those from Chain A are colored in red. D: Stereoview of the superposition of Nup145N(443–605) dimers from two different crystal forms. Nup145N(443–605) forms an identical dimer in both tetragonal and hexagonal crystal forms. All structural superpositions were carried out using SSM. The A and B chains of tetragonal form are shown as gray and green ribbons, respectively. Similarly, A and B chains of the hexagonal forms are shown in yellow and wheat, respectively.
Figure 2
A: Analytical gel filtration profile of Nup145N(443–605) at concentrations of 11.8 (black), 22.7 (blue), and 46.0mg/ml (red) with elution times 21.5, 21.3, and 21.1min, respectively. B: Comparison of the merged experimental SAXS profile (red) of Nup145N(443–605) with SAXS profiles computed by IMP for the crystallographic dimer (blue) and monomer (green) structures (inset shows the SAXS profiles across the entire measured resolution range). C: The shape of Nup145N(443–605) represented as mesh derived from the experimental SAXS profile.
Figure 3
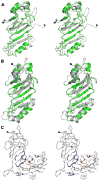
A: Stereoview of Nup145N(443–605, green) superposed on a representative NMR structure of Nup116 (PDB Code 2AIV; grey). B: Stereoview of the Nup145N(443–605, chain B; green) superposed on Nup98-N (grey) from the Nup98-N:C-terminal fragment complex (PDB Code 2Q5Y). C: Stereoview of the dimeric association of Nup98-N in the Nup98-N:C-terminal fragment complex. The ordered C-terminal fragment tri-peptides are shown in magenta. Residues contributing to the dimer interface from the A and C chains are colored blue and orange, respectively. The N- and C-terminal residues are shown on the cartoons as blue and red spheres, respectively.
Similar articles
- Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Sampathkumar P, Kim SJ, Manglicmot D, Bain KT, Gilmore J, Gheyi T, Phillips J, Pieper U, Fernandez-Martinez J, Franke JD, Matsui T, Tsuruta H, Atwell S, Thompson DA, Emtage JS, Wasserman SR, Rout MP, Sali A, Sauder JM, Almo SC, Burley SK. Sampathkumar P, et al. Proteins. 2012 Aug;80(8):2110-6. doi: 10.1002/prot.24102. Epub 2012 Jun 4. Proteins. 2012. PMID: 22544723 Free PMC article. - Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Sampathkumar P, Gheyi T, Miller SA, Bain KT, Dickey M, Bonanno JB, Kim SJ, Phillips J, Pieper U, Fernandez-Martinez J, Franke JD, Martel A, Tsuruta H, Atwell S, Thompson DA, Emtage JS, Wasserman SR, Rout MP, Sali A, Sauder JM, Burley SK. Sampathkumar P, et al. Proteins. 2011 May;79(5):1672-7. doi: 10.1002/prot.22973. Epub 2011 Mar 1. Proteins. 2011. PMID: 21365675 Free PMC article. - Nuclear pores. Architecture of the nuclear pore complex coat.
Stuwe T, Correia AR, Lin DH, Paduch M, Lu VT, Kossiakoff AA, Hoelz A. Stuwe T, et al. Science. 2015 Mar 6;347(6226):1148-52. doi: 10.1126/science.aaa4136. Epub 2015 Feb 12. Science. 2015. PMID: 25745173 Free PMC article. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review. - Nuclear pore complexes: round the bend?
Antonin W, Mattaj IW. Antonin W, et al. Nat Cell Biol. 2005 Jan;7(1):10-2. doi: 10.1038/ncb0105-10. Nat Cell Biol. 2005. PMID: 15632943 Review. No abstract available.
Cited by
- Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Sampathkumar P, Kim SJ, Manglicmot D, Bain KT, Gilmore J, Gheyi T, Phillips J, Pieper U, Fernandez-Martinez J, Franke JD, Matsui T, Tsuruta H, Atwell S, Thompson DA, Emtage JS, Wasserman SR, Rout MP, Sali A, Sauder JM, Almo SC, Burley SK. Sampathkumar P, et al. Proteins. 2012 Aug;80(8):2110-6. doi: 10.1002/prot.24102. Epub 2012 Jun 4. Proteins. 2012. PMID: 22544723 Free PMC article. - An ancient autoproteolytic domain found in GAIN, ZU5 and Nucleoporin98.
Liao Y, Pei J, Cheng H, Grishin NV. Liao Y, et al. J Mol Biol. 2014 Dec 12;426(24):3935-3945. doi: 10.1016/j.jmb.2014.10.011. Epub 2014 Oct 20. J Mol Biol. 2014. PMID: 25451782 Free PMC article. - The yeast nuclear pore complex and transport through it.
Aitchison JD, Rout MP. Aitchison JD, et al. Genetics. 2012 Mar;190(3):855-83. doi: 10.1534/genetics.111.127803. Genetics. 2012. PMID: 22419078 Free PMC article. - Evolutionary divergence of the nuclear pore complex from fungi to metazoans.
Chopra K, Bawaria S, Chauhan R. Chopra K, et al. Protein Sci. 2019 Mar;28(3):571-586. doi: 10.1002/pro.3558. Epub 2018 Dec 24. Protein Sci. 2019. PMID: 30488506 Free PMC article. - Integrative structure and functional anatomy of a nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, Chemmama IE, Pellarin R, Echeverria I, Shivaraju M, Chaudhury AS, Wang J, Williams R, Unruh JR, Greenberg CH, Jacobs EY, Yu Z, de la Cruz MJ, Mironska R, Stokes DL, Aitchison JD, Jarrold MF, Gerton JL, Ludtke SJ, Akey CW, Chait BT, Sali A, Rout MP. Kim SJ, et al. Nature. 2018 Mar 22;555(7697):475-482. doi: 10.1038/nature26003. Epub 2018 Mar 14. Nature. 2018. PMID: 29539637 Free PMC article.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang WH, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang WZ, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 RR022220-01/RR/NCRR NIH HHS/United States
- U54 GM074945/GM/NIGMS NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- R01 GM062427-11/GM/NIGMS NIH HHS/United States
- U54 GM074945-06/GM/NIGMS NIH HHS/United States
- R01 GM062427/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases