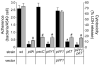Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin - PubMed (original) (raw)
Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin
Ryan W Heiniger et al. Cell Microbiol. 2010 Aug.
Abstract
Tissue damage predisposes humans to life-threatening disseminating infection by the opportunistic pathogen Pseudomonas aeruginosa. Bacterial adherence to host tissue is a critical first step in this infection process. It is well established that P. aeruginosa attachment to host cells involves type IV pili (TFP), which are retractile surface fibres. The molecular details of attachment and the identity of the bacterial adhesin and host receptor remain controversial. Using a mucosal epithelium model system derived from primary human tissue, we show that the pilus-associated protein PilY1 is required for bacterial adherence. We establish that P. aeruginosa preferentially binds to exposed basolateral host cell surfaces, providing a mechanistic explanation for opportunistic infection of damaged tissue. Further, we demonstrate that invasion and fulminant infection of intact host tissue requires the coordinated and mutually dependent action of multiple bacterial factors, including pilus fibre retraction and the host cell intoxication system, termed type III secretion. Our findings offer new and important insights into the complex interactions between a pathogen and its human host and provide compelling evidence that PilY1 serves as the principal P. aeruginosa adhesin for human tissue and that it specifically recognizes a host receptor localized or enriched on basolateral epithelial cell surfaces.
Figures
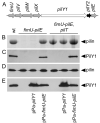
Fig. 1. PilY1 is conditionally required for TFP production
Surface TFP were visualized by (A) transmission electron microscopy and (B) immunofluorescence microscopy. Pilus fibers (thin) are visible on the surface of the wild type (wt) strain and a non-retractile pilT mutant. In contrast, a pilY1 mutant is devoid of TFP and indistinguishable from a non-piliated control strain (pilA). Wild type levels of TFP are restored in a pilY1 mutant complemented with plasmid-expressed pilY1 (pPa-pilY1). Pilus fibers are also restored in a pilY1 mutant when pilT is inactivated (pilY1, pilT), indicating PilY1 is not an essential TFP biogenesis factor. Strain genotype is indicated in each panel. For electron microscopy, scale bars equal 500 nm. Thick fibers visible on the wt strain and pilA, pilT and pilY1 mutants are flagella. For immunofluorescence, bacterial cells were labeled with DAPI (blue) and TFP were labeled with pilin-specific antibody, Alexa Red 594-conjugated goat anti-rabbit secondary antibody (red) and bacteria were imaged at 100X magnification.
Fig. 2. PilY1 is conditionally required for TFP production and co-localizes with sheared surface fibers
(A) Coomassie Blue-stained SDS-PAGE gel showing the relative abundance of recovered pilin in sheared pilus fractions. (B) Immunoblot of pilus fractions probed with PilY1-specific antiserum. (C-E) Immunoblots of whole bacterial cell lysates separated by SDS-PAGE and probed with pilin-specific (C), PilY1-specific (D) or PilT-specific (E) antiserum.
Fig. 3. Localization of PilY1 to the pilus fraction requires proteins encoded by the _pilY1_-associated operon
(A) Organization of the _pilY1_-associated operon. Pilin-like genes are shaded grey. (B) Coomassie Blue-stained SDS-PAGE gel showing the relative abundance of recovered pilin in pilus fractions. (C) Immunoblot of surface pilus fractions probed with PilY1-specific antiserum. (D-E) Whole bacterial cell lysates separated by SDS-PAGE and probed with either pilin-specific (D) or PilY1-specific (E) antiserum by immunoblotting.
Fig. 4. Retractile TFP and pilus-associated PilY1 are necessary for productive interactions with A549 epithelial cells
Bacterial adherence (black bars; mean +/− SEM; _n_=5) and cytotoxicity (grey bars; mean +/− SEM; _n_=3) to A549 cells is shown. Bacterial adherence was determined based on the average number of bacteria bound per A549 cell. Adherence to A549 cells was significantly reduced (*; p<0.001) in the non-pilated pilA and pilY1 mutants and in the piliated non-retractile pilT and pilY1, pilT mutants compared to wild type. Cytotoxicity was determined based on the percentage (%) of lactate dehydrogenase (LDH) released from A549 cells following bacterial infection relative to the amount of LDH released following cell lysis with 0.25% Triton X-100. The ability to elicit a cytotoxic effect was significantly reduced (#; p<0.001) in the non-adherent strains and in a T3S mutant (pscC) compared to wild type.
Fig. 5. Invasive infection of Human Airway Epithelium (HAE) cultures requires retractile TFP and Type III Secretion
Histological cross-sections of HAE cultures inoculated with wild type (wt), pilA, pilT and pscC strains. Representative images taken at 3, 6, and 12 hours post-infection are shown. Wild type P. aeruginosa rarely adhered to the ciliated mucosal surfaces but local infection foci could be detected after 3 hours. After breaching the mucosal barrier, the wild type strain interacted efficiently with the basolateral surfaces of ciliated cells and the underlying basal epithelial cells. Bacterial adherence was associated with host cell rounding and detachment. By 12 hours, the wild type infection appeared to spread between cells to encompass the entire HAE culture. In contrast, the non-piliated pilA mutant, the piliated non-retractile pilT mutant and the T3S mutant (pscC) did not interact with the epithelium or cause invasive infection. However, unlike the pilA mutant, the pilT and pscC mutants retained the ability to interact with shed or extruding epithelial cells (bottom right panels).
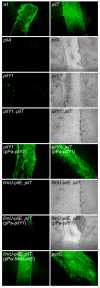
Fig. 6. TFP and pilus-associated PilY1 are required for adherence of P. aeruginosa to injured HAE cultures
Injured HAE cultures were imaged (en face) by fluorescence microscopy 45 minutes after inoculation with bacterial strains expressing GFP. The wild type (wt) strain, the piliated pilT mutant and a non-cytotoxic T3S mutant (pscC) attached efficiently to the injured tissue but showed limited interaction with the adjacent intact epithelium. The non-piliated (pilY1) and piliated (pilY1, pilT) pilY1 mutants did not adhere to the injured tissue and were indistinguishable from the non-piliated control mutant (pilA). Adherence was restored for both the non-piliated and piliated pilY1 mutants following complementation with plasmid-expressed PilY1 (pPa-pilY1). The piliated fimU-pilE, pilT mutant was unable to adhere to the damaged tissue; complementation with the entire fimU operon (pPa-fimU-pilE) but not pilY1 alone (pPa-pilY1) restored adherence. For non-adherent strains, tissue damage was confirmed by examining the HAE cultures by both fluorescence and light microscopy.
Fig. 7. PilY1 is required for adherence to exposed basal epithelial cells of injured HAE cultures
Histological sections of HAE cultures show the presence of bacteria on newly exposed basal epithelial cells after injury. The wild type strain (wt), the piliated pilT mutant and a non-cytotoxic T3S mutant (pscC) showed robust adherence, while the non-piliated (pilY1) and piliated (pilY1, pilT) PilY1-lacking strains were defective for binding to basal epithelial cells at the injury site. Adherence was restored for both the non-piliated and piliated pilY1 mutants following complementation with plasmid-expressed pilY1 (pPa-pilY1). Only strains which retained twitching motility and cytotoxicity (wt and the complemented pilY1 mutant) were able to further penetrate the exposed basal cell layers.
Fig. 8. Model of events leading to invasive infection of the human airway epithelium by P. aeruginosa
The three different phases of epithelial infection (apical interaction, penetration and dissemination) and the P. aeruginosa factors required for each phase are indicated. (A) Apical interaction. Productive interactions between P. aeruginosa and the intact mucosal epithelium is a relatively rare event exploiting transiently exposed basolateral surfaces during cell extrusion. This event is dependent on bacterial adherence mediated by TFP and TFP-associated PilY1. (B) Penetration. Subsequent penetration of the mucosal barrier requires retractile TFP and T3S. PilT-dependent pilus retraction may facilitate contact-dependent T3S and TFP-mediated bacterial motility. Cytotoxicity mediated by T3S causes additional tissue damage and disruption of host cell junctions and exposure of additional basolateral host receptors. (C) Dissemination. Following the formation of a focal infection, PilY1-mediated adherence, retractile TFP and T3S act synergistically to cause fulminant infection and dissemination into deeper tissue.
Similar articles
- Pseudomonas aeruginosa minor pilins prime type IVa pilus assembly and promote surface display of the PilY1 adhesin.
Nguyen Y, Sugiman-Marangos S, Harvey H, Bell SD, Charlton CL, Junop MS, Burrows LL. Nguyen Y, et al. J Biol Chem. 2015 Jan 2;290(1):601-11. doi: 10.1074/jbc.M114.616904. Epub 2014 Nov 11. J Biol Chem. 2015. PMID: 25389296 Free PMC article. - The interaction of Pseudomonas aeruginosa PAK with human and animal respiratory tract cell lines.
Hambrook J, Titball R, Lindsay C. Hambrook J, et al. FEMS Microbiol Lett. 2004 Sep 1;238(1):49-55. doi: 10.1016/j.femsle.2004.07.016. FEMS Microbiol Lett. 2004. PMID: 15336402 - Global Regulatory Pathways Converge To Control Expression of Pseudomonas aeruginosa Type IV Pili.
Coggan KA, Higgs MG, Brutinel ED, Marden JN, Intile PJ, Winther-Larsen HC, Koomey M, Yahr TL, Wolfgang MC. Coggan KA, et al. mBio. 2022 Feb 22;13(1):e0369621. doi: 10.1128/mbio.03696-21. Epub 2022 Jan 25. mBio. 2022. PMID: 35073734 Free PMC article. - The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa--a review.
Hahn HP. Hahn HP. Gene. 1997 Jun 11;192(1):99-108. doi: 10.1016/s0378-1119(97)00116-9. Gene. 1997. PMID: 9224879 Review. - Adhesins and receptors of Pseudomonas aeruginosa associated with infection of the respiratory tract.
Prince A. Prince A. Microb Pathog. 1992 Oct;13(4):251-60. doi: 10.1016/0882-4010(92)90035-m. Microb Pathog. 1992. PMID: 1363702 Review.
Cited by
- Role of Mfa5 in Expression of Mfa1 Fimbriae in Porphyromonas gingivalis.
Hasegawa Y, Iijima Y, Persson K, Nagano K, Yoshida Y, Lamont RJ, Kikuchi T, Mitani A, Yoshimura F. Hasegawa Y, et al. J Dent Res. 2016 Oct;95(11):1291-7. doi: 10.1177/0022034516655083. Epub 2016 Jun 20. J Dent Res. 2016. PMID: 27323953 Free PMC article. - Mechanism of assembly of type 4 filaments: everything you always wanted to know (but were afraid to ask).
Pelicic V. Pelicic V. Microbiology (Reading). 2023 Mar;169(3):001311. doi: 10.1099/mic.0.001311. Microbiology (Reading). 2023. PMID: 36947586 Free PMC article. Review. - The Pseudomonas aeruginosa AlgZR two-component system coordinates multiple phenotypes.
Okkotsu Y, Little AS, Schurr MJ. Okkotsu Y, et al. Front Cell Infect Microbiol. 2014 Jun 20;4:82. doi: 10.3389/fcimb.2014.00082. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 24999454 Free PMC article. Review. - Current concepts on Pseudomonas aeruginosa interaction with human airway epithelium.
Muggeo A, Coraux C, Guillard T. Muggeo A, et al. PLoS Pathog. 2023 Mar 30;19(3):e1011221. doi: 10.1371/journal.ppat.1011221. eCollection 2023 Mar. PLoS Pathog. 2023. PMID: 36996043 Free PMC article. Review. - The Type IVa Pilus Machinery Is Recruited to Sites of Future Cell Division.
Carter T, Buensuceso RN, Tammam S, Lamers RP, Harvey H, Howell PL, Burrows LL. Carter T, et al. mBio. 2017 Jan 31;8(1):e02103-16. doi: 10.1128/mBio.02103-16. mBio. 2017. PMID: 28143978 Free PMC article.
References
- Alm RA, Hallinan JP, Watson AA, Mattick JS. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. - PubMed
- Alm RA, Mattick JS. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL080098/HL/NHLBI NIH HHS/United States
- R01 AI069116/AI/NIAID NIH HHS/United States
- HL084934/HL/NHLBI NIH HHS/United States
- R21 HL080098/HL/NHLBI NIH HHS/United States
- P50 HL084934/HL/NHLBI NIH HHS/United States
- AI069116/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources